You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003912_00453
You are here: Home > Sequence: MGYG000003912_00453
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000435635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000435635 | |||||||||||
| CAZyme ID | MGYG000003912_00453 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 144021; End: 146744 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 73 | 881 | 3.3e-227 | 0.9956958393113343 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15979 | Glyco_hydro_115 | 9.93e-164 | 228 | 569 | 1 | 334 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| pfam17829 | GH115_C | 2.84e-24 | 752 | 897 | 15 | 171 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
| TIGR01564 | S_layer_MJ | 0.001 | 59 | 197 | 421 | 569 | S-layer protein, MJ0822 family. This model represents one of several families of proteins associated with the formation of prokaryotic S-layers. Members of this family are found in archaeal species, including Pyrococcus horikoshii (split into two tandem reading frames), Methanococcus jannaschii, and related species. Some local similarity can be found to other S-layer protein families. [Cell envelope, Surface structures] |
| pfam05124 | S_layer_C | 0.002 | 126 | 197 | 149 | 219 | S-layer like family, C-terminal region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALK86808.1 | 0.0 | 75 | 902 | 33 | 967 |
| EDV07546.1 | 0.0 | 75 | 902 | 32 | 966 |
| AYD49386.1 | 1.89e-264 | 43 | 902 | 3 | 966 |
| APZ45930.1 | 9.61e-258 | 44 | 902 | 5 | 960 |
| AUC19792.1 | 9.61e-258 | 44 | 902 | 5 | 960 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BY3_A | 2.00e-154 | 74 | 884 | 19 | 773 | Anovel family GH115 4-O-Methyl-alpha-glucuronidase, BtGH115A, with specificity for decorated arabinogalactans [Bacteroides thetaiotaomicron VPI-5482] |
| 4C90_A | 1.05e-135 | 32 | 894 | 3 | 844 | Evidencethat GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
| 7PXQ_A | 1.65e-119 | 75 | 892 | 16 | 826 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 7PUG_A | 3.30e-119 | 59 | 892 | 1 | 827 | ChainA, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
| 4ZMH_A | 3.34e-115 | 77 | 896 | 16 | 930 | Crystalstructure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
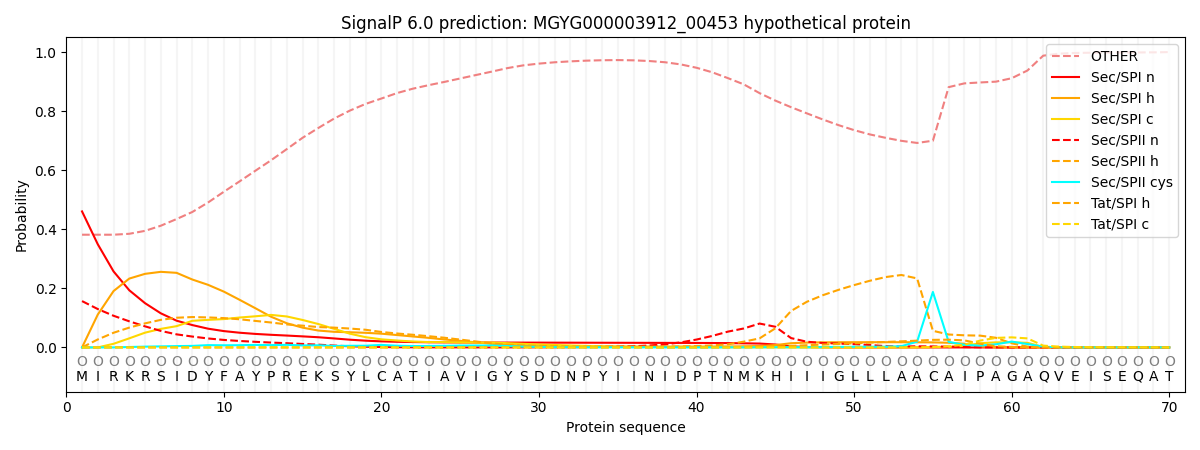
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.389460 | 0.322491 | 0.272531 | 0.011721 | 0.002750 | 0.001040 |
