You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004037_01792
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471;
|
| CAZyme ID |
MGYG000004037_01792
|
| CAZy Family |
CE19 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000004037 |
2199612 |
MAG |
United Kingdom |
Europe |
|
| Gene Location |
Start: 7418;
End: 8824
Strand: +
|
No EC number prediction in MGYG000004037_01792.
| Family |
Start |
End |
Evalue |
family coverage |
| CE19 |
132 |
462 |
8.8e-115 |
0.9879518072289156 |
MGYG000004037_01792 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6GOC_A
|
1.50e-169 |
33 |
464 |
25 |
455 |
ChainA, DUF3826 domain-containing protein [Bacteroides thetaiotaomicron] |
This protein is predicted as SP
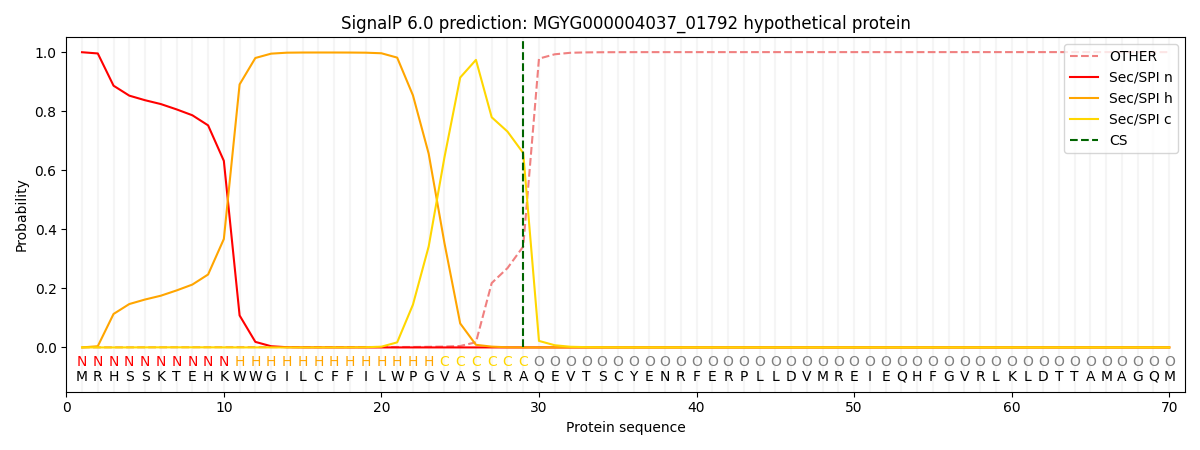
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.001276
|
0.997722
|
0.000358
|
0.000198
|
0.000194
|
0.000204
|
There is no transmembrane helices in MGYG000004037_01792.

