You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004045_01424
You are here: Home > Sequence: MGYG000004045_01424
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
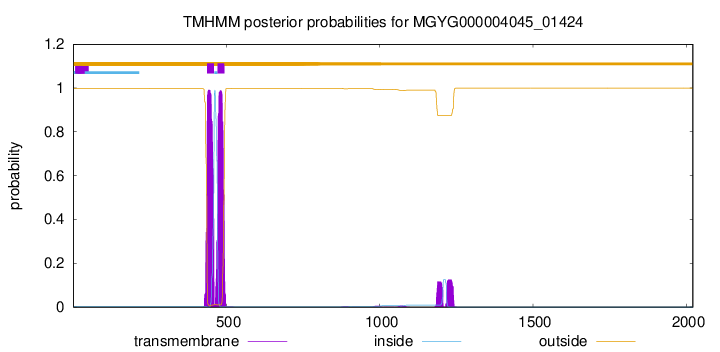
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus; | |||||||||||
| CAZyme ID | MGYG000004045_01424 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11021; End: 17089 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13402 | LT_TF-like | 1.69e-47 | 1788 | 1905 | 1 | 116 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG3953 | SLT | 9.96e-36 | 1792 | 1901 | 19 | 135 | SLT domain protein [Mobilome: prophages, transposons]. |
| COG1196 | Smc | 6.94e-14 | 55 | 401 | 687 | 1025 | Chromosome segregation ATPase [Cell cycle control, cell division, chromosome partitioning]. |
| TIGR02168 | SMC_prok_B | 9.74e-14 | 3 | 401 | 700 | 1048 | chromosome segregation protein SMC, common bacterial type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. This family represents the SMC protein of most bacteria. The smc gene is often associated with scpB (TIGR00281) and scpA genes, where scp stands for segregation and condensation protein. SMC was shown (in Caulobacter crescentus) to be induced early in S phase but present and bound to DNA throughout the cell cycle. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
| COG1196 | Smc | 1.03e-10 | 38 | 437 | 173 | 514 | Chromosome segregation ATPase [Cell cycle control, cell division, chromosome partitioning]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHC53060.1 | 0.0 | 1 | 2022 | 1 | 2022 |
| QTP20427.1 | 0.0 | 1 | 2022 | 1 | 2022 |
| QTD66356.1 | 0.0 | 1 | 2022 | 1 | 2021 |
| AXQ19242.1 | 0.0 | 1 | 2022 | 1 | 2017 |
| AOG25514.1 | 0.0 | 1 | 2022 | 1 | 2021 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P45931 | 7.97e-13 | 1796 | 1962 | 1372 | 1539 | Uncharacterized protein YqbO OS=Bacillus subtilis (strain 168) OX=224308 GN=yqbO PE=1 SV=2 |
| P54334 | 1.70e-12 | 1702 | 1909 | 1041 | 1235 | Phage-like element PBSX protein XkdO OS=Bacillus subtilis (strain 168) OX=224308 GN=xkdO PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000064 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |