You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004501_01300
You are here: Home > Sequence: MGYG000004501_01300
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900760665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900760665 | |||||||||||
| CAZyme ID | MGYG000004501_01300 | |||||||||||
| CAZy Family | GH148 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15642; End: 18824 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH148 | 435 | 585 | 1.4e-38 | 0.9868421052631579 |
| GH148 | 643 | 760 | 2.3e-17 | 0.7697368421052632 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 0.004 | 430 | 510 | 4 | 81 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44571.1 | 1.22e-147 | 30 | 1059 | 33 | 1114 |
| AHF94074.1 | 5.41e-140 | 153 | 1058 | 157 | 1094 |
| AVM44690.1 | 4.06e-121 | 150 | 1060 | 167 | 1104 |
| AVM44197.1 | 4.69e-120 | 21 | 1060 | 27 | 1095 |
| AVM43772.1 | 6.16e-120 | 13 | 1059 | 10 | 1108 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3PZ9_A | 1.56e-07 | 431 | 585 | 24 | 188 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 2.42e-07 | 431 | 585 | 10 | 174 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
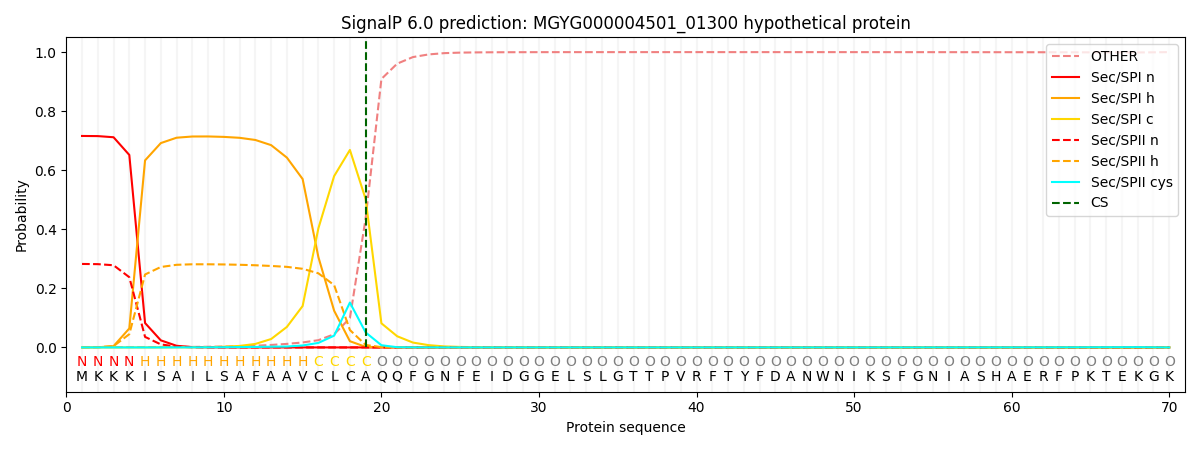
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002687 | 0.708528 | 0.288061 | 0.000230 | 0.000236 | 0.000232 |
