

| Basic Information | |
|---|---|
| Species | Chlamydomonas reinhardtii |
| Cazyme ID | Cre10.g450500.t1.2 |
| Family | CBM20 |
| Protein Properties | Length: 762 Molecular Weight: 72820.2 Isoelectric Point: 3.6084 |
| Chromosome | Chromosome/Scaffold: 10 Start: 4258498 End: 4264436 |
| Description | catalytics;carbohydrate kinases;phosphoglucan, water dikinases |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM20 | 57 | 136 | 1.6e-21 |
| KMPYRVNFGETLRLVGSSPLLGDWNADKGLQLEWSEGDVWCADVPIDAGHYEFKCVIYNAQSNQVSRWEDGGNRVLNITE | |||
| Full Sequence |
|---|
| Protein Sequence Length: 762 Download |
| MMRSFACNQK CSRTANAPRA FTLASPSAPA PFVVRRRVAL VVRNDAGAST KPTLSIKMPY 60 RVNFGETLRL VGSSPLLGDW NADKGLQLEW SEGDVWCADV PIDAGHYEFK CVIYNAQSNQ 120 VSRWEDGGNR VLNITEEPGA WAVDCLWGAT TDMTQAFVPA PPPPAPEPVA EVAPVAEAPA 180 SEASASGAAA IPAVEPITAT TSTVSETAAV AIETAAAAIE TAAAVIDTAP AVTTEATAAT 240 PATVTAVPTA FAVVIQPAGL QPATEPTTEP TTEPATTETA NTEPATELVI AAALNAVAET 300 VDAPPAAAEA EAPAVASVPA AMELGVVPPA EAAVPPVEAA AAAIAAAATE AAAVEAAAVE 360 AAAVEAAAVE AAAAEAAAVE AAAIEAASSE AAAVEAAAVE AAAVEAAAAV AEAAAVEAAS 420 SEAAAVEAAA VVAAAPVMVE VAEVAAETVA TAQVVEPPVV EAVAEAAVES AMAAAAVAPA 480 MDTASVTEAA VLETAAAVTA AAAALEAAEP AATADAVVKG ADTAAAVAEE SLKSATAAAG 540 VEEELLPTFL ALEEAGAAKA AEEQAAADVA AAAAAEAAAA SAAASTSGTG GDAASGSGAG 600 PIALLLATAA AGAAAAWVLL APGGIDTSAL GSGASSALGG ASEAVNKTLL QSSLAGAQVR 660 AQLQSQVTAL AGQVSSVSAQ VASLQPLVQE QVSSLSDGMA AAATGAAASL GSLSATSVAV 720 VQSTAEGVGG AVTGALGGLA AGMDGLQAVA AEAVEQVARQ G* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
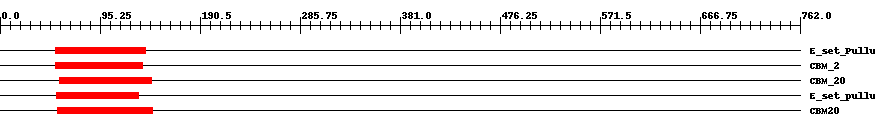 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd02860 | E_set_Pullulanase | 4.0e-12 | 53 | 139 | 89 | + Early set domain associated with the catalytic domain of pullulanase (also called dextrinase and alpha-dextrin endo-1,6-alpha glucosidase). E or "early" set domains are associated with the catalytic domain of pullulanase at either the N-terminal or C-terminal end, and in a few instances at both ends. Pullulanase is an enzyme with activity similar to that of isoamylase; it cleaves 1,6-alpha-glucosidic linkages in pullulan, amylopectin, and glycogen, and in alpha-and beta-amylase limit-dextrins of amylopectin and glycogen. The E set domain of pullulanase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. | ||
| smart01065 | CBM_2 | 2.0e-14 | 53 | 136 | 88 | + Starch binding domain. | ||
| pfam00686 | CBM_20 | 2.0e-15 | 57 | 144 | 92 | + Starch binding domain. | ||
| cd02861 | E_set_pullulanase_like | 3.0e-16 | 54 | 132 | 80 | + Early set domain associated with the catalytic domain of pullulanase-like proteins. E or "early" set domains are associated with the catalytic domain of pullulanase at either the N-terminal or C-terminal end, and in a few instances at both ends. Pullulanase (also called dextrinase or alpha-dextrin endo-1,6-alpha glucosidase) is an enzyme with action similar to that of isoamylase; it cleaves 1,6-alpha-glucosidic linkages in pullulan, amylopectin, and glycogen, and in alpha-and beta-amylase limit-dextrins of amylopectin and glycogen. The E set domain of pullulanase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. | ||
| cd05467 | CBM20 | 3.0e-19 | 55 | 145 | 95 | + The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. | ||
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_001696233.1 | 0.0000001 | 36 | 130 | 17 | 104 | hypothetical protein CHLREDRAFT_182288 [Chlamydomonas reinhardtii] |
| RefSeq | XP_001768545.1 | 0.00000008 | 52 | 132 | 101 | 183 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | ZP_02692213.1 | 0.0000003 | 337 | 409 | 166 | 236 | signal recognition particle-docking protein FtsY [Epulopiscium sp. 'N.t. morphotype B'] |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
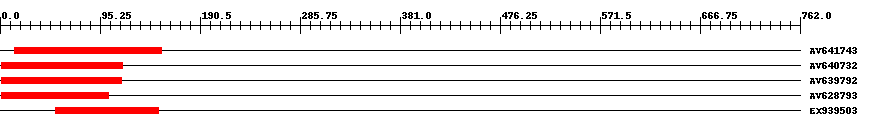 | ||||
| Hit | Length | Start | End | EValue |
| AV641743 | 141 | 14 | 154 | 0 |
| AV640732 | 117 | 1 | 117 | 0 |
| AV639792 | 116 | 1 | 116 | 0 |
| AV628793 | 103 | 1 | 103 | 0 |
| EX939503 | 101 | 53 | 151 | 0.000000000002 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |