

| Basic Information | |
|---|---|
| Species | Volvox carteri |
| Cazyme ID | Vocar20000733m |
| Family | CBM20 |
| Protein Properties | Length: 1907 Molecular Weight: 190090 Isoelectric Point: 5.7593 |
| Chromosome | Chromosome/Scaffold: 19 Start: 698886 End: 707466 |
| Description | Protein phosphatase 2C family protein |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM20 | 86 | 168 | 6.1e-21 |
| KVRLCFQHQVPFGQVLKVVGSGPKLGDWQPDHAQTLQWSDGHLWTADVTLEPGSYELKCVVAVGDSIIEWERGANRILQVPSG | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1907 Download |
| MTFARPSQGL AGNNGRNIYR VVRLGCHFRP RFLSCQHLGL QQRCNNEGQQ LTKSWHSNKN 60 AEARSSLQRC QAASITTQAR EARTIKVRLC FQHQVPFGQV LKVVGSGPKL GDWQPDHAQT 120 LQWSDGHLWT ADVTLEPGSY ELKCVVAVGD SIIEWERGAN RILQVPSGVR GIRVDGSWSR 180 TADNRITVTE PIAAASVAAR QKRAAAARLL AKALAEVNAA AGPTATSLAQ RRKTAAAGAA 240 DESAPSKGQA AGGAGDSAAA EAEAARKEDD ADSLRELRAL LSPSVSSSKP PASAGVVATA 300 SLTPPGGGNG GSGGKRPNGD GVQVGVVVVG GVASGNVERS QGSGGKEVQQ QQAKDELGTC 360 TEQQQEQELK GQDSNQQPAG GGRVSGRGSW RRELLSALVA NGEEQPPVQR QEDQQAGRES 420 ASSASGGLAI GGGGKSSSYS SSNIAFGDGN GLPGGLSTAS QQQQQRERER EQQPRQQLAG 480 GEGAAAATAT GDIAQSIREA AAARGEPLGA AREVPTGSAD GAVAPVVAGE AERAPVTQPA 540 TSQQASARTA TAAAEAAAAP AADAAAAEAL PSTASPPPAV PVVSVVRSLS QNNRPPAAAT 600 AAASEPSGGD PSERSSVGSG GPGGNVRFAN LLSRLGGVVG ANLQRGPAAK TESKTSGVVT 660 GADGKEDLTR AVPTPVPSAG SSSSSGGGGS KARRAASWRQ LLDDGGEEER RVRDGPSAAN 720 VQQQQQSAAS QSQEVVKGGD KERNPEPKVV QRKVEEEGKE EEGFTARFTQ LLTSRLTGQP 780 LRYPAAASGT PTPAAQPPAT PAAAAAAATA VGGFDRIQAH EEKEGEKVKS VEVQEGQEEV 840 QEEKVGKSAT RQSQEIPSEV QVTELLAAPS TPPGRDNRPP PPPAVAAAVA AAAAGGVAAT 900 TTSAPSGAAA SNRLISSSSS TSCNSGRNVG GGRRDSSSSS SKAEAGDAAS SPSSSDRDMG 960 TTSSRESHPR NGEVESVNDP GLERLIGAWD SAWSQGRVKQ PAATAEVPRP NGGGGGSSSG 1020 SSNDGNGAAG TVNGGATATG TSVVKPYVVV AFSGTGSSSA SANDRAQGQQ ESAVAASAAG 1080 KLANGGVAAP PGGAAGGSGE SLYSIAERLT GGSGPLAAQL CSSDESSPGI TVRMGAMAAM 1140 GVDRQGSFDN RSSYLSQSLL AESLNLNGGG GGASGQDPSS TALLRQLRAA NAANAGAADC 1200 TGSTANGGGA PAAAPVPNLP ATPAPPGILS SVDEDEDGVV MPPRLPATPP PPSYLSGAAG 1260 SGSGPAAAAP VPPLPATPVS SSILSGSLNG VGAAAEDLLR RASDADAVLN MLRGNLPLTP 1320 VARGTRGGGG GPLSMAEDGE EDEQNSSSTN NNNKDARNGR SRRGRRGAGT RLIPVLPIPG 1380 VTGDSQIDPL TAARAVAADA AQALDKTRAT LQALQQGAGL GLTAEGLSES AARLLQQLPK 1440 LDALGGGLAA GGGQDGGEAN GNAGGAGVTA PGLPPAVSQL ATSLVSGLGT VGSSLTDQLR 1500 KAATVLPSGS AASTAEAANG RGSDASANGN GSAGAAGLTP MAADGGGAGG GLLSGLLGSL 1560 VPGSTPLPPG CVRLVAGAHM IPHVDKVDKG GEDAYFISRV GLGGVGVADG VSGWADEGID 1620 PAEYPRTLMR FAADAFEAAR GTMSAPDIIR YAQYRTYLKG SSTVCMALMK PGKRLEVANV 1680 GDSGVRILRN GKVIFGTEAQ QHAFNMPYQL SHPNNVEDPD SADDADVHIV DVQEGDVVML 1740 ATDGLFDNVF DEEIEQVVSQ QLRELAAAGR GRAPMTAAVT VNGGGEAAAA AAAASSSGSG 1800 GTGASGRSGK GLLSQSFSAA AAAAAAAAGA GAAGGGGNSY RPEDAARVAR ALAERAHLHA 1860 RNPTQRTPWS VTSSQQPNFM WAKFFAKGGG KMDDCTVLVA FVCGPE* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
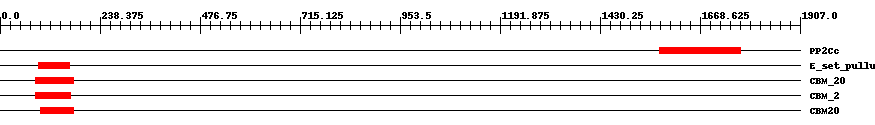 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| smart00332 | PP2Cc | 2.0e-11 | 1571 | 1764 | 227 | + Serine/threonine phosphatases, family 2C, catalytic domain. The protein architecture and deduced catalytic mechanism of PP2C phosphatases are similar to the PP1, PP2A, PP2B family of protein Ser/Thr phosphatases, with which PP2C shares no sequence similarity. | ||
| cd02861 | E_set_pullulanase_like | 1.0e-11 | 92 | 165 | 77 | + Early set domain associated with the catalytic domain of pullulanase-like proteins. E or "early" set domains are associated with the catalytic domain of pullulanase at either the N-terminal or C-terminal end, and in a few instances at both ends. Pullulanase (also called dextrinase or alpha-dextrin endo-1,6-alpha glucosidase) is an enzyme with action similar to that of isoamylase; it cleaves 1,6-alpha-glucosidic linkages in pullulan, amylopectin, and glycogen, and in alpha-and beta-amylase limit-dextrins of amylopectin and glycogen. The E set domain of pullulanase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. | ||
| pfam00686 | CBM_20 | 3.0e-14 | 84 | 175 | 96 | + Starch binding domain. | ||
| smart01065 | CBM_2 | 2.0e-14 | 85 | 167 | 88 | + Starch binding domain. | ||
| cd05467 | CBM20 | 2.0e-16 | 97 | 175 | 83 | + The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0005975 | carbohydrate metabolic process |
| GO:2001070 | starch binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | NP_565696.1 | 1e-39 | 1566 | 1761 | 40 | 231 | catalytic [Arabidopsis thaliana] |
| RefSeq | XP_001697518.1 | 4e-39 | 100 | 217 | 4 | 121 | predicted protein [Chlamydomonas reinhardtii] |
| RefSeq | XP_001697518.1 | 0 | 1100 | 1903 | 818 | 1572 | predicted protein [Chlamydomonas reinhardtii] |
| RefSeq | XP_002273518.1 | 1e-39 | 1572 | 1760 | 58 | 243 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002516354.1 | 1e-39 | 1572 | 1795 | 47 | 266 | protein phosphatase 2c, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1kum_A | 0.005 | 97 | 178 | 19 | 107 | A Chain A, The Structure Of Cbescii Cela Gh9 Module |
| PDB | 1kul_A | 0.005 | 97 | 178 | 19 | 107 | A Chain A, The Structure Of Cbescii Cela Gh9 Module |
| PDB | 1acz_A | 0.005 | 97 | 178 | 19 | 107 | A Chain A, Glucoamylase, Granular Starch-Binding Domain Complex With Cyclodextrin, Nmr, 5 Structures |
| PDB | 1ac0_A | 0.005 | 97 | 178 | 19 | 107 | A Chain A, Glucoamylase, Granular Starch-Binding Domain Complex With Cyclodextrin, Nmr, Minimized Average Structure |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
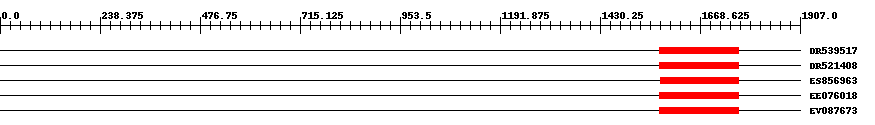 | ||||
| Hit | Length | Start | End | EValue |
| DR539517 | 189 | 1572 | 1760 | 3e-35 |
| DR521408 | 189 | 1572 | 1760 | 4e-35 |
| ES856963 | 187 | 1574 | 1760 | 5e-35 |
| EE076018 | 189 | 1572 | 1760 | 1e-34 |
| EV087673 | 190 | 1572 | 1761 | 5e-34 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |