

| Basic Information | |
|---|---|
| Species | Sorghum bicolor |
| Cazyme ID | Sb02g041935.1 |
| Family | GT1 |
| Protein Properties | Length: 469 Molecular Weight: 49961.8 Isoelectric Point: 6.7308 |
| Chromosome | Chromosome/Scaffold: 2 Start: 75663261 End: 75664667 |
| Description | UDP-Glycosyltransferase superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 36 | 450 | 1.5e-37 |
| LPVHFAAPEPHLREARARLHGWDGTLPAAVRFRALDVPPHESPSPDPSSPFPAHMLPLFEAYCGGARAPLAALLAELSATHRRVVVLHDRMAAFAAAEAA RLPNADALGVHCLAASYNVGWADREHALLRRHGLVFHPPDACATQEFVALARRMGQERRRAPGAGMVVNTCRALEGDFLDALQGIPSSDGQRLFAVGPLS PVLLPLPGAGASSGTSRRHECLDWLDAQPASSVLYVSFGTTSSLRPEQVRELAAALRDSGARFVWVLRDADRADLRSGGEGDAAAETLRAAAASELGPGA SRAGVVVTGWAPQLEILAHGATAAFMSHCGWNSTVESLSHGKPILAWPMHSDQPWDAELVCKYLRAGVLVRPWERRHDVIPAAAIRDAIDRVMASDEGAE IRRRAAALGEAVRGA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 469 Download |
| MEMESAAVAI VTVPFPAQGH LNQLLHLSLL LAARGLPVHF AAPEPHLREA RARLHGWDGT 60 LPAAVRFRAL DVPPHESPSP DPSSPFPAHM LPLFEAYCGG ARAPLAALLA ELSATHRRVV 120 VLHDRMAAFA AAEAARLPNA DALGVHCLAA SYNVGWADRE HALLRRHGLV FHPPDACATQ 180 EFVALARRMG QERRRAPGAG MVVNTCRALE GDFLDALQGI PSSDGQRLFA VGPLSPVLLP 240 LPGAGASSGT SRRHECLDWL DAQPASSVLY VSFGTTSSLR PEQVRELAAA LRDSGARFVW 300 VLRDADRADL RSGGEGDAAA ETLRAAAASE LGPGASRAGV VVTGWAPQLE ILAHGATAAF 360 MSHCGWNSTV ESLSHGKPIL AWPMHSDQPW DAELVCKYLR AGVLVRPWER RHDVIPAAAI 420 RDAIDRVMAS DEGAEIRRRA AALGEAVRGA VAEGGSSRQD LDELVAYVT |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02554 | PLN02554 | 3.0e-31 | 174 | 411 | 238 | + UDP-glycosyltransferase family protein | ||
| PLN02992 | PLN02992 | 1.0e-32 | 200 | 436 | 244 | + coniferyl-alcohol glucosyltransferase | ||
| PLN02448 | PLN02448 | 1.0e-35 | 248 | 469 | 224 | + UDP-glycosyltransferase family protein | ||
| PLN03007 | PLN03007 | 1.0e-35 | 254 | 405 | 152 | + UDP-glucosyltransferase family protein | ||
| PLN00164 | PLN00164 | 6.0e-36 | 193 | 395 | 204 | + glucosyltransferase; Provisional | ||
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ACD03254.1 | 0 | 7 | 469 | 10 | 464 | UDP-glycosyltransferase UGT93B9 [Avena strigosa] |
| GenBank | EAY95124.1 | 0 | 7 | 469 | 8 | 462 | hypothetical protein OsI_16941 [Oryza sativa Indica Group] |
| GenBank | EEE67749.1 | 0 | 12 | 469 | 14 | 461 | hypothetical protein OsJ_25448 [Oryza sativa Japonica Group] |
| RefSeq | NP_001060532.1 | 0 | 12 | 469 | 14 | 477 | Os07g0660500 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002463331.1 | 0 | 1 | 469 | 1 | 469 | hypothetical protein SORBIDRAFT_02g041935 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2pq6_A | 9.94922e-44 | 10 | 468 | 11 | 477 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt85h2- Insights Into The Structural Basis Of A Multifunctional (Iso) Flavonoid Glycosyltransferase |
| PDB | 2acv_B | 1.99965e-42 | 10 | 469 | 12 | 462 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2acv_A | 1.99965e-42 | 10 | 469 | 12 | 462 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2acw_B | 1.99965e-42 | 10 | 469 | 12 | 462 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2acw_A | 1.99965e-42 | 10 | 469 | 12 | 462 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| cytokinins-O-glucoside biosynthesis | RXN-4735 | EC-2.4.1.215 | cis-zeatin O-β-D-glucosyltransferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
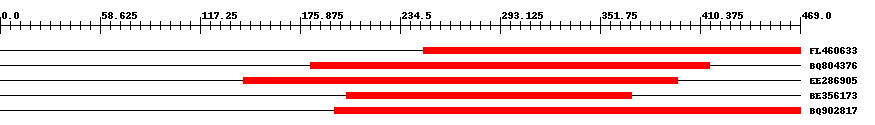 | ||||
| Hit | Length | Start | End | EValue |
| FL460633 | 222 | 248 | 469 | 0 |
| BQ804376 | 238 | 182 | 416 | 0 |
| EE286905 | 255 | 143 | 397 | 0 |
| BE356173 | 168 | 203 | 370 | 0 |
| BQ902817 | 275 | 196 | 469 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |