

| Basic Information | |
|---|---|
| Species | Sorghum bicolor |
| Cazyme ID | Sb08g002010.1 |
| Family | GT8 |
| Protein Properties | Length: 655 Molecular Weight: 74803.1 Isoelectric Point: 8.5084 |
| Chromosome | Chromosome/Scaffold: 8 Start: 2032779 End: 2037117 |
| Description | galacturonosyltransferase 7 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT8 | 323 | 627 | 0 |
| CLNMRLTVEYFKSRSSHMDQLNEQELESPTFHHYVIFSKNVLAASTTINSAVMNSQNSDHIVFHLFTDAQNFYAMKHWFDRNSYLEATVHVTNIEDNQNL SKDMHSLEMQQLWPAEEFRVTIRNHSEPSQRQMKTEYISIFGHSHFLLPDLLPSLNRVVVLDDDLIVQKDLSSLWNLDMGGKVIGAVQFCEVRLGQLKPY MADHNVNANSCVWLSGLNVVELDKWRDMGITSLYDQSFQKLRKDRLKSQRFQALPASLLAFQDLVYPLEDSWVQSGLGHDYGISHVDIEKAATLHYNGVM KPWLD | |||
| Full Sequence |
|---|
| Protein Sequence Length: 655 Download |
| MKGHQHSSSP LLPTPPPSKR RCSGLAAAVP ALVVCSTLLP LVYLLVLHRP AAGYGSDDRA 60 AVVISTELAG VGARGKRHLE NGGAMKHKLL KDVSKKVSGS NGIPAERSTR SKSNKDHAIK 120 SKAKLKGAFS LIGLNNDTFK SKGPRTPKRY QLKDLSWRSK DTTVNGKENY GQEAVHEEYT 180 KSCEHEYGSY CLWSTEHREV MKDAIVKRLK DQLFLARAHY PSIAKLKQQE RFTRELKQNI 240 QEHERMLSDT ITDADLPPFF AKKLEKMEHT IERAKSCEVG CSNVERKLRQ LLDITEDEAY 300 FHTRQSAFLY HLGVQTTPKT HHCLNMRLTV EYFKSRSSHM DQLNEQELES PTFHHYVIFS 360 KNVLAASTTI NSAVMNSQNS DHIVFHLFTD AQNFYAMKHW FDRNSYLEAT VHVTNIEDNQ 420 NLSKDMHSLE MQQLWPAEEF RVTIRNHSEP SQRQMKTEYI SIFGHSHFLL PDLLPSLNRV 480 VVLDDDLIVQ KDLSSLWNLD MGGKVIGAVQ FCEVRLGQLK PYMADHNVNA NSCVWLSGLN 540 VVELDKWRDM GITSLYDQSF QKLRKDRLKS QRFQALPASL LAFQDLVYPL EDSWVQSGLG 600 HDYGISHVDI EKAATLHYNG VMKPWLDLGI HDYKSYWREY MTNGEKFMTE CNIH* 660 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
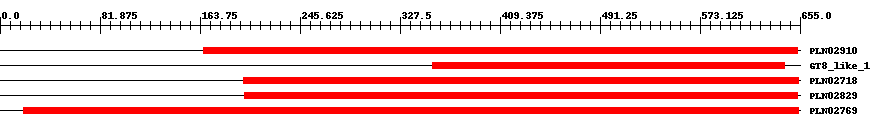 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02910 | PLN02910 | 3.0e-103 | 167 | 653 | 502 | + polygalacturonate 4-alpha-galacturonosyltransferase | ||
| cd06429 | GT8_like_1 | 2.0e-104 | 354 | 642 | 289 | + GT8_like_1 represents a subfamily of GT8 with unknown function. A subfamily of glycosyltransferase family 8 with unknown function: Glycosyltransferase family 8 comprises enzymes with a number of known activities; lipopolysaccharide galactosyltransferase lipopolysaccharide glucosyltransferase 1, glycogenin glucosyltransferase and inositol 1-alpha-galactosyltransferase. It is classified as a retaining glycosyltransferase, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. | ||
| PLN02718 | PLN02718 | 3.0e-105 | 199 | 654 | 465 | + Probable galacturonosyltransferase | ||
| PLN02829 | PLN02829 | 2.0e-115 | 200 | 653 | 467 | + Probable galacturonosyltransferase | ||
| PLN02769 | PLN02769 | 0 | 19 | 654 | 636 | + Probable galacturonosyltransferase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0016757 | transferase activity, transferring glycosyl groups |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABA95691.2 | 0 | 71 | 654 | 1 | 596 | Glycosyl transferase family 8 protein, expressed [Oryza sativa (japonica cultivar-group)] |
| GenBank | EEC67573.1 | 0 | 1 | 654 | 1 | 642 | hypothetical protein OsI_34919 [Oryza sativa Indica Group] |
| GenBank | EEC68769.1 | 0 | 19 | 654 | 17 | 659 | hypothetical protein OsI_37298 [Oryza sativa Indica Group] |
| RefSeq | NP_001065619.1 | 0 | 1 | 654 | 1 | 642 | Os11g0124900 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002441763.1 | 0 | 1 | 654 | 1 | 654 | hypothetical protein SORBIDRAFT_08g002010 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1ga8_A | 0.001 | 455 | 625 | 74 | 252 | A Chain A, Structure Of The Thermostable Gh51 Alpha-L-Arabinofuranosidase From Thermotoga Petrophila Rku-1 |
| PDB | 1g9r_A | 0.001 | 455 | 625 | 74 | 252 | A Chain A, Crystal Structure Of Galactosyltransferase Lgtc In Complex With Mn And Udp-2f-Galactose |
| PDB | 1ss9_A | 0.002 | 455 | 548 | 74 | 165 | A Chain A, Crystal Structural Analysis Of Active Site Mutant Q189e Of Lgtc |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| homogalacturonan biosynthesis | 2.4.1.43-RXN | EC-2.4.1.43 | polygalacturonate 4-α-galacturonosyltransferase |