

| Basic Information | |
|---|---|
| Species | Mimulus guttatus |
| Cazyme ID | mgv1a000878m |
| Family | GT35 |
| Protein Properties | Length: 953 Molecular Weight: 107479 Isoelectric Point: 5.1954 |
| Chromosome | Chromosome/Scaffold: 11 Start: 164507 End: 170537 |
| Description | Glycosyl transferase, family 35 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT35 | 164 | 947 | 0 |
| ALEKLGHTLETVASQEPDAALGNGGLGRLASCFLDSLATLNYPAWGYGLRYKYGLFKQQLTKDGQEEVAENWLENGNPWEIVRNDVCYPVKFSGKVVEGS DGKKRWIGGEDIVAVAYDVPIPGYKTKTTINLRLWSTKVPSDQFDLHAFNAGEHTKACEAQANAEKICYVLYPGDESVEGKILRLKQQYTLCSASLQDII ARFERRSGGDVRWEDFPEKVAVQMNDTHPTLCIPELMRILMDLKGMSWDEAWRITKRTVAYTNHTVLPEALEKWSYDLMQRLLPRHVEIIEKIDEQLIED IVSEYGTLNPEMLEKKLATMRILENFDLPASIADLFAKPEESPVDETSEEVKSKDEVTVTEKDEQLDGEETQKNKAVHKEPAYIPPKMVRMANLCVVGGH TVNGVAEIHSEIVKQEVFNDFFQLWPEKFQNKTNGVTPRRWIQYCNPDLSAVITKWIGSNDWVLNTDKLAELRKFADNEDLQREWRAAKKSNKIKLVSFL KEKTGYSVNPDAMFDIQVKRIHEYKRQLLNILGIVYRYKKMKEMTADERKANFVPRVCIFGGKAFSTYVQAKRIVKFITDVGATINHDPDIGDLLKVIFV PDYNVSVAELLIPASELSQHISTAGMEASGTSNMKFSMNGCILIGTLDGANVEIREEVGDDNFFLFGAQAHEIAPLRNERAAGEFVPDERFEEVKKFVRS GAFGAYNYDEMIGSLEGNEGFGRADYFLVGKDFPSYIECQDKVDIAYRDQKRWTKMSILNTAGSYKFSSDRTIREYANDIWNIE | |||
| Full Sequence |
|---|
| Protein Sequence Length: 953 Download |
| MAASSTGAAV TAYSLCSSNA RLIDFASRDR SSKVLLLARV KPSFCVRCVS SEPKQRVRDP 60 IAEEGVLSNL SALSPHAASI ASSIKYHAEF TPMFSPQNFE PPKAFFAAAQ SVRDALIINW 120 NATNDFYEKM NVKQAYYLSM EFLQGRALLN AIGNLELSGE YADALEKLGH TLETVASQEP 180 DAALGNGGLG RLASCFLDSL ATLNYPAWGY GLRYKYGLFK QQLTKDGQEE VAENWLENGN 240 PWEIVRNDVC YPVKFSGKVV EGSDGKKRWI GGEDIVAVAY DVPIPGYKTK TTINLRLWST 300 KVPSDQFDLH AFNAGEHTKA CEAQANAEKI CYVLYPGDES VEGKILRLKQ QYTLCSASLQ 360 DIIARFERRS GGDVRWEDFP EKVAVQMNDT HPTLCIPELM RILMDLKGMS WDEAWRITKR 420 TVAYTNHTVL PEALEKWSYD LMQRLLPRHV EIIEKIDEQL IEDIVSEYGT LNPEMLEKKL 480 ATMRILENFD LPASIADLFA KPEESPVDET SEEVKSKDEV TVTEKDEQLD GEETQKNKAV 540 HKEPAYIPPK MVRMANLCVV GGHTVNGVAE IHSEIVKQEV FNDFFQLWPE KFQNKTNGVT 600 PRRWIQYCNP DLSAVITKWI GSNDWVLNTD KLAELRKFAD NEDLQREWRA AKKSNKIKLV 660 SFLKEKTGYS VNPDAMFDIQ VKRIHEYKRQ LLNILGIVYR YKKMKEMTAD ERKANFVPRV 720 CIFGGKAFST YVQAKRIVKF ITDVGATINH DPDIGDLLKV IFVPDYNVSV AELLIPASEL 780 SQHISTAGME ASGTSNMKFS MNGCILIGTL DGANVEIREE VGDDNFFLFG AQAHEIAPLR 840 NERAAGEFVP DERFEEVKKF VRSGAFGAYN YDEMIGSLEG NEGFGRADYF LVGKDFPSYI 900 ECQDKVDIAY RDQKRWTKMS ILNTAGSYKF SSDRTIREYA NDIWNIEPLE IN* 960 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
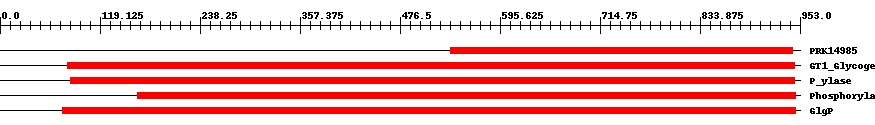 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK14985 | PRK14985 | 5.0e-145 | 537 | 944 | 417 | + maltodextrin phosphorylase; Provisional | ||
| cd04300 | GT1_Glycogen_Phosphorylase | 0 | 81 | 946 | 875 | + This is a family of oligosaccharide phosphorylases. It includes yeast and mammalian glycogen phosphorylases, plant starch/glucan phosphorylase, as well as the maltodextrin phosphorylases of bacteria. The members of this family catalyze the breakdown of oligosaccharides into glucose-1-phosphate units. They are important allosteric enzymes in carbohydrate metabolism. The allosteric control mechanisms of yeast and mammalian members of this family are different from that of bacterial members. The members of this family belong to the GT-B structural superfamily of glycoslytransferases, which have characteristic N- and C-terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. | ||
| TIGR02093 | P_ylase | 0 | 84 | 946 | 871 | + glycogen/starch/alpha-glucan phosphorylases. This family consists of phosphorylases. Members use phosphate to break alpha 1,4 linkages between pairs of glucose residues at the end of long glucose polymers, releasing alpha-D-glucose 1-phosphate. The nomenclature convention is to preface the name according to the natural substrate, as in glycogen phosphorylase, starch phosphorylase, maltodextrin phosphorylase, etc. Name differences among these substrates reflect differences in patterns of branching with alpha 1,6 linkages. Members include allosterically regulated and unregulated forms. A related family, TIGR02094, contains examples known to act well on particularly small alpha 1,4 glucans, as may be found after import from exogenous sources [Energy metabolism, Biosynthesis and degradation of polysaccharides]. | ||
| pfam00343 | Phosphorylase | 0 | 164 | 948 | 792 | + Carbohydrate phosphorylase. The members of this family catalyze the formation of glucose 1-phosphate from one of the following polyglucoses; glycogen, starch, glucan or maltodextrin. | ||
| COG0058 | GlgP | 0 | 74 | 948 | 880 | + Glucan phosphorylase [Carbohydrate transport and metabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004645 | phosphorylase activity |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAA36612.1 | 0 | 3 | 951 | 2 | 965 | unnamed protein product [Solanum tuberosum] |
| EMBL | CBI22291.1 | 0 | 12 | 951 | 15 | 981 | unnamed protein product [Vitis vinifera] |
| Swiss-Prot | P04045 | 0 | 3 | 951 | 2 | 965 | PHSL1_SOLTU RecName: Full=Alpha-1,4 glucan phosphorylase L-1 isozyme, chloroplastic/amyloplastic; AltName: Full=Starch phosphorylase L-1; Flags: Precursor |
| Swiss-Prot | P27598 | 0 | 19 | 949 | 20 | 952 | PHSL_IPOBA RecName: Full=Alpha-1,4 glucan phosphorylase L isozyme, chloroplastic/amyloplastic; AltName: Full=Starch phosphorylase L; Flags: Precursor |
| RefSeq | XP_002279075.1 | 0 | 12 | 951 | 15 | 957 | PREDICTED: similar to Alpha-1,4 glucan phosphorylase L-1 isozyme, chloroplastic/amyloplastic [Vitis vinifera] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1ygp_B | 0 | 87 | 949 | 51 | 878 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 1ygp_A | 0 | 87 | 949 | 51 | 878 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 3cem_B | 0 | 76 | 948 | 1 | 807 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 3cem_A | 0 | 76 | 948 | 1 | 807 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| PDB | 3cej_B | 0 | 76 | 948 | 1 | 807 | A Chain A, Phosphorylated Form Of Yeast Glycogen Phosphorylase With Phosphate Bound In The Active Site. |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| HO778303 | 864 | 104 | 951 | 0 |
| HO797178 | 399 | 553 | 951 | 0 |
| HO613954 | 511 | 441 | 951 | 0 |
| HO418036 | 586 | 290 | 872 | 0 |
| HO778303 | 121 | 1 | 106 | 0.000000000005 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |