CAZyme Information
You are here: Home >
Sequence: NP_212393.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
Signalp annotations |
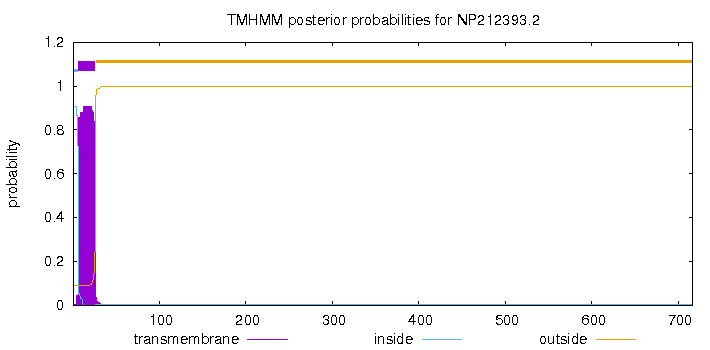
TMHMM annotations |
PSSpred annotations |
LipoP annotations
Basic Information help
| Species | Borrelia burgdorferi B31 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZyme ID | NP_212393.2 | |||||||||||||
| CAZy Family | GH23; | |||||||||||||
| CAZyme Description | transglycosylase SLT domain-containing protein | |||||||||||||
| CAZyme Property |
|
|||||||||||||
| Genome Property |
|
|||||||||||||
| Gene Location | Start: 270488; End: 272641 Strand: - | |||||||||||||
| View CDS | CDS sequence | |||||||||||||
| UniProt ID | O51274; | |||||||||||||
| InterPro Annotation | O51274; | |||||||||||||
Genomic Context If it does not show, please wait or refresh. help
Enzyme Prediction help
| metacyc | EC |
|---|---|
| RXN0-5190; |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | Start | End | Domain Description |
|---|---|---|---|---|---|
| pfam01464 | SLT | 2.31e-34 | 559 | 671 |
Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems.
|
| COG0741 | MltE | 6.62e-30 | 551 | 701 |
Soluble lytic murein transglycosylase and related regulatory proteins (some contain LysM/invasin domains) [Cell wall/membrane/envelope biogenesis].
|
| cd00254 | LT_GEWL | 4.30e-24 | 571 | 690 |
Lytic Transglycosylase (LT) and Goose Egg White Lysozyme (GEWL) domain. Members include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as, the eukaryotic "goose-type" lysozymes (GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue.
|
CAZyme Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIU79502.1| GH23 | 0.0e+00 | 1 | 717 | 1 | 717 |
| AAC66641.2| GH23 | 0.0e+00 | 1 | 717 | 1 | 717 |
| ADQ30537.1| GH23 | 0.0e+00 | 1 | 717 | 1 | 717 |
| AGS66278.1| GH23 | 0.0e+00 | 1 | 717 | 16 | 732 |
| ADQ29271.1| GH23 | 0.0e+00 | 1 | 717 | 1 | 717 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YP4_A | 2.1e-17 | 535 | 698 | 436 | 599 |
Chain A, Snm-deletion Mutant (lytic Transglycosylase)
|
| 5MPQ_A | 2.1e-17 | 535 | 698 | 396 | 559 |
Chain A, Bulgecin A: The Key To A Broad-spectrum Inhibitor That Targets Lytic Transglycosylases
|
| 4YIM_B | 2.1e-17 | 535 | 698 | 406 | 569 |
Chain B, Structural Characterization Of The Lytic Transglycosylase Dependent Mechanism Of Cytotoxic Peptidoglycan Fragment Release.
|
| 1SLY_A | 7.8e-17 | 551 | 694 | 449 | 598 |
Chain A, Complex Of The 70-Kda Soluble Lytic Transglycosylase With Bulgecin A
|
| 1QSA_A | 3.0e-16 | 551 | 694 | 449 | 598 |
Chain A, Crystal Structure Of The 70 Kda Soluble Lytic Transglycosylase Slt70 From Escherichia Coli At 1.65 Angstroms Resolution
|
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P39434|SLT_SALTY | 5.2e-16 | 512 | 694 | 428 | 625 |
Soluble lytic murein transglycosylase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) GN=slt PE=3 SV=2
|
| sp|P0AGC4|SLT_ECO57 | 2.6e-15 | 551 | 694 | 476 | 625 |
Soluble lytic murein transglycosylase OS=Escherichia coli O157:H7 GN=slt PE=3 SV=1
|
| sp|P0AGC3|SLT_ECOLI | 2.6e-15 | 551 | 694 | 476 | 625 |
Soluble lytic murein transglycosylase OS=Escherichia coli (strain K12) GN=slt PE=1 SV=1
|
| sp|B0BSH0|MLTC_ACTPJ | 4.7e-09 | 555 | 687 | 197 | 354 |
Membrane-bound lytic murein transglycosylase C OS=Actinobacillus pleuropneumoniae serotype 3 (strain JL03) GN=mltC PE=3 SV=1
|
| sp|B3GYW8|MLTC_ACTP7 | 4.7e-09 | 555 | 687 | 197 | 354 |
Membrane-bound lytic murein transglycosylase C OS=Actinobacillus pleuropneumoniae serotype 7 (strain AP76) GN=mltC PE=3 SV=1
|
PSSpred Annotations download dat.ss data help
LipoP Annotations download full data without filtering help
| class | score | margin | cleavage |
|---|---|---|---|
| SpII | 5.89085 | 4.32419 | 23-24 |