

| Basic Information | |
|---|---|
| Species | Populus trichocarpa |
| Cazyme ID | Potri.009G087900.1 |
| Family | CBM22 |
| Protein Properties | Length: 580 Molecular Weight: 65460.9 Isoelectric Point: 8.5014 |
| Chromosome | Chromosome/Scaffold: 09 Start: 8152253 End: 8156047 |
| Description | Glycosyl hydrolase family 10 protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM22 | 27 | 142 | 4.4e-21 |
| IIVNPELNDGLRAWSTFGDAKTEHRESNGNKYVVAHSRNNPYGSMSQKLYLKKNHLYTFSAWVQVSEGNVQVTAIFKTDSGFKKAGSVFAEPKCWSMLKG GLTVDASGPAELYFES | |||
| GH10 | 202 | 466 | 0 |
| NKLRFPFGCAINKNILSNTAYQDWFTSRFGVTAFEDEMKWYSTEATRGQVDYSVPDAMMAFAKQHNIAVRGHNVIWDDPKYQSGWVNSLSPNDFRTAVQA RVGSVMTRYRGRLLAWDVVNENMHFSFVESKLGQNASSVIYNSAGKTDGLTTLFLNEYDTIEKSGEGAASPAKYLQKLKEIQSFPGNANLRMGIGLESHF TIPNLPYMRASLDTLASANVPIWLTEVDVQGNPAQQAQYLEQILREGYSYPKIAGIVMWSAWKPQ | |||
| Full Sequence |
|---|
| Protein Sequence Length: 580 Download |
| MPCPMITRSA LRQCLENPLS PQYNGGIIVN PELNDGLRAW STFGDAKTEH RESNGNKYVV 60 AHSRNNPYGS MSQKLYLKKN HLYTFSAWVQ VSEGNVQVTA IFKTDSGFKK AGSVFAEPKC 120 WSMLKGGLTV DASGPAELYF ESNNTSVEIW VDSISLQPFT EKEWRSHQDQ SIERTRKEKV 180 RIQAIDEQGN PLSNATISIK QNKLRFPFGC AINKNILSNT AYQDWFTSRF GVTAFEDEMK 240 WYSTEATRGQ VDYSVPDAMM AFAKQHNIAV RGHNVIWDDP KYQSGWVNSL SPNDFRTAVQ 300 ARVGSVMTRY RGRLLAWDVV NENMHFSFVE SKLGQNASSV IYNSAGKTDG LTTLFLNEYD 360 TIEKSGEGAA SPAKYLQKLK EIQSFPGNAN LRMGIGLESH FTIPNLPYMR ASLDTLASAN 420 VPIWLTEVDV QGNPAQQAQY LEQILREGYS YPKIAGIVMW SAWKPQGCYR MCLTDNNFKN 480 LATGDVVDKL LHEWGGSLMG MTDANGFFEA SLSHGDYNEG VWIKTLHSGG FYIPHAHMEE 540 HIKKREKDKI CLPNNQREGR INDHKRSQKR RRFCLAIWT* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam02018 | CBM_4_9 | 6.0e-6 | 34 | 146 | 123 | + Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. | ||
| COG3693 | XynA | 3.0e-27 | 211 | 479 | 300 | + Beta-1,4-xylanase [Carbohydrate transport and metabolism] | ||
| pfam00331 | Glyco_hydro_10 | 2.0e-46 | 205 | 460 | 274 | + Glycosyl hydrolase family 10. | ||
| smart00633 | Glyco_10 | 9.0e-70 | 239 | 487 | 273 | + Glycosyl hydrolase family 10. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAN10199.1 | 0 | 9 | 517 | 35 | 544 | endoxylanase [Carica papaya] |
| RefSeq | XP_002264556.1 | 0 | 9 | 521 | 115 | 624 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002264605.1 | 0 | 9 | 540 | 30 | 564 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002313180.1 | 0 | 13 | 529 | 2 | 514 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002532418.1 | 0 | 9 | 518 | 26 | 536 | Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1nq6_A | 4e-29 | 208 | 479 | 15 | 292 | A Chain A, Crystal Structure Of The Catalytic Domain Of Xylanase A From Streptomyces Halstedii Jm8 |
| PDB | 1ta3_B | 1e-22 | 208 | 460 | 17 | 268 | B Chain B, Crystal Structure Of Xylanase (Gh10) In Complex With Inhibitor (Xip) |
| PDB | 1v0n_A | 8e-22 | 208 | 460 | 16 | 266 | B Chain B, Crystal Structure Of Xylanase (Gh10) In Complex With Inhibitor (Xip) |
| PDB | 1v0m_A | 8e-22 | 208 | 460 | 16 | 266 | B Chain B, Crystal Structure Of Xylanase (Gh10) In Complex With Inhibitor (Xip) |
| PDB | 1v0l_A | 8e-22 | 208 | 460 | 16 | 266 | B Chain B, Crystal Structure Of Xylanase (Gh10) In Complex With Inhibitor (Xip) |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| (1,4)-β-xylan degradation | 3.2.1.8-RXN | EC-3.2.1.8 | endo-1,4-β-xylanase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
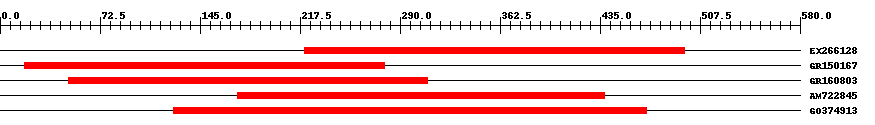 | ||||
| Hit | Length | Start | End | EValue |
| EX266128 | 278 | 221 | 496 | 0 |
| GR150167 | 263 | 18 | 279 | 0 |
| GR160803 | 262 | 50 | 310 | 0 |
| AM722845 | 268 | 172 | 438 | 0 |
| GO374913 | 346 | 126 | 469 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |