

| Basic Information | |
|---|---|
| Species | Mimulus guttatus |
| Cazyme ID | mgv1a023615m |
| Family | GH1 |
| Protein Properties | Length: 431 Molecular Weight: 48584.7 Isoelectric Point: 6.4446 |
| Chromosome | Chromosome/Scaffold: 48 Start: 31826 End: 34798 |
| Description | beta glucosidase 14 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH1 | 308 | 403 | 7.1e-27 |
| EWLFVVPKGFYELLVHTYKVYDNPLIFVTENGLDVFVSFSVGARVKGYFVWSLLDNYEWAEGYTVRFGMIYVDYGNGLTRYPKSSAIWFMNFLSRK | |||
| GH1 | 11 | 304 | 0 |
| RHDFDTEFVFGAATSAYQIEGGYKEGGKAFSIWDTFSITKPAKISDGSNGCLAIDHYNKFKEDIDLMKKLGLDAYRLSFAWTRILPGGRLSGGVNREGVK FYNDVIDYALSKGIQPYVTIFHWDIPTFLQEEYGGFASPKIVQDFAEYAEVCFFEFGDRVKHWITINESWTYTHHGYISGDFPPNHGSYSTGAESTNGAP TIEKKIHRCGSGVDQTVNSLSNPGTEPYEVAHNLILAHAAAVEVYRRDYQEAQKGIIGVTNVSKWFIPLKDTKEDKAAAARAVDFMWGWFIAPI | |||
| Full Sequence |
|---|
| Protein Sequence Length: 431 Download |
| MAGSDNSNIS RHDFDTEFVF GAATSAYQIE GGYKEGGKAF SIWDTFSITK PAKISDGSNG 60 CLAIDHYNKF KEDIDLMKKL GLDAYRLSFA WTRILPGGRL SGGVNREGVK FYNDVIDYAL 120 SKGIQPYVTI FHWDIPTFLQ EEYGGFASPK IVQDFAEYAE VCFFEFGDRV KHWITINESW 180 TYTHHGYISG DFPPNHGSYS TGAESTNGAP TIEKKIHRCG SGVDQTVNSL SNPGTEPYEV 240 AHNLILAHAA AVEVYRRDYQ EAQKGIIGVT NVSKWFIPLK DTKEDKAAAA RAVDFMWGWF 300 IAPIGGSEWL FVVPKGFYEL LVHTYKVYDN PLIFVTENGL DVFVSFSVGA RVKGYFVWSL 360 LDNYEWAEGY TVRFGMIYVD YGNGLTRYPK SSAIWFMNFL SRKTLLYDKR QVEEKIESVN 420 KNPAKRPRQG * |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG2723 | BglB | 1.0e-17 | 297 | 396 | 125 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| PLN02849 | PLN02849 | 6.0e-76 | 2 | 304 | 303 | + beta-glucosidase | ||
| COG2723 | BglB | 5.0e-77 | 14 | 304 | 292 | + Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism] | ||
| TIGR03356 | BGL | 1.0e-108 | 18 | 396 | 466 | + beta-galactosidase. | ||
| pfam00232 | Glyco_hydro_1 | 7.0e-109 | 14 | 403 | 467 | + Glycosyl hydrolase family 1. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAF03675.1 | 0 | 4 | 309 | 12 | 312 | AF149311_1 raucaffricine-O-beta-D-glucosidase [Rauvolfia serpentina] |
| GenBank | AAF03675.1 | 1e-25 | 301 | 429 | 384 | 540 | AF149311_1 raucaffricine-O-beta-D-glucosidase [Rauvolfia serpentina] |
| GenBank | AAL93619.1 | 0 | 2 | 309 | 26 | 328 | beta-glucosidase [Olea europaea subsp. europaea] |
| GenBank | AAL93619.1 | 6e-25 | 302 | 414 | 395 | 539 | beta-glucosidase [Olea europaea subsp. europaea] |
| RefSeq | XP_002448029.1 | 0 | 9 | 402 | 32 | 485 | hypothetical protein SORBIDRAFT_06g019860 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 4atl_B | 0 | 4 | 304 | 12 | 307 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 4atl_B | 8e-27 | 301 | 398 | 384 | 509 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 4atl_A | 0 | 4 | 304 | 12 | 307 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 4atl_A | 8e-27 | 301 | 398 | 384 | 509 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 4atd_B | 0 | 4 | 304 | 12 | 307 | A Chain A, Crystal Structure Of Native Raucaffricine Glucosidase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
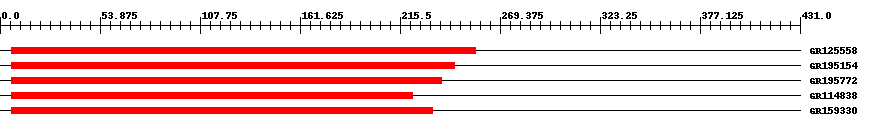 | ||||
| Hit | Length | Start | End | EValue |
| GR125558 | 253 | 6 | 256 | 0 |
| GR195154 | 244 | 6 | 245 | 0 |
| GR195772 | 237 | 6 | 238 | 0 |
| GR114838 | 227 | 6 | 222 | 0 |
| GR159330 | 230 | 6 | 233 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |