

| Basic Information | |
|---|---|
| Species | Ricinus communis |
| Cazyme ID | 29729.m002282 |
| Family | GH85 |
| Protein Properties | Length: 687 Molecular Weight: 76703.7 Isoelectric Point: 6.7799 |
| Chromosome | Chromosome/Scaffold: 29729 Start: 80926 End: 84294 |
| Description | Glycosyl hydrolase family 85 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH85 | 73 | 355 | 0 |
| MQGGYVDDKWVQGGNNKSAYAIWHWYLIDVFVYFSHNLVTLPPPCWTNTAHRHGVKVLGTFITEGSDGTETCNKLLATKESAHMYAERLAELAADLGFDG WLMNIEVELEAKQIPNLKEFVSHLTQIMHSTIPGSLVIWYDSVTVNGRLIYQNQLNENNKPFFDICDGIFANYWWAKDYPKNSAVVAGDRKFDVYMGVDV FGRGTYGGGEWNTNVALDVCKKADVSAAIFAPGWVYETKQPPDFQTAQNKWWSLVEKSCGVVKSYPNTLPFYSNFDQGHGYHF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 687 Download |
| MSLTNEAQSS TLDPPPFDPL QPSIPVSYPL KTLKELESRS YFKSFHYSFN KSSVSLKSSG 60 LDNRPRILVC HDMQGGYVDD KWVQGGNNKS AYAIWHWYLI DVFVYFSHNL VTLPPPCWTN 120 TAHRHGVKVL GTFITEGSDG TETCNKLLAT KESAHMYAER LAELAADLGF DGWLMNIEVE 180 LEAKQIPNLK EFVSHLTQIM HSTIPGSLVI WYDSVTVNGR LIYQNQLNEN NKPFFDICDG 240 IFANYWWAKD YPKNSAVVAG DRKFDVYMGV DVFGRGTYGG GEWNTNVALD VCKKADVSAA 300 IFAPGWVYET KQPPDFQTAQ NKWWSLVEKS CGVVKSYPNT LPFYSNFDQG HGYHFSVEGG 360 QVSNAPWNNI SSQGLQPFLE FNKNQTTDTI QVLADFKEAS YSGGANITFK GTLKDHNDFT 420 ARLFQGRLLL GELPLHMTYS VKSDGDSQIG LCLYFSSTLN KRTSVFIAPC GKSQFSNEFS 480 KVIVPHRVDK PEMAPGWVIQ ESSIDMNGYT LTEIHALCYR SKPEHGKLRS EYISDRHDNT 540 TGPSPSEYFA VLGHITIKNS KENPVFPASS SWLVAGQCIK WISGSQGSKK LSIKISWKLK 600 DGSTSQFSKF NIYVEKLGKN AGRNSDGRIE GIQEFIGVAC VETFYVSCLS IPCSTSSVKF 660 IIQMCGIDGT CQKLVDSPLF LLDVEGQ |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
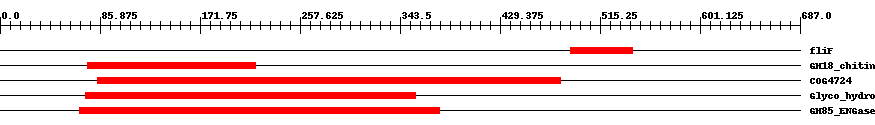 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK12800 | fliF | 0.009 | 490 | 543 | 54 | + flagellar MS-ring protein; Reviewed | ||
| cd00598 | GH18_chitinase-like | 0.0001 | 75 | 219 | 164 | + The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. | ||
| COG4724 | COG4724 | 8.0e-41 | 84 | 481 | 431 | + Endo-beta-N-acetylglucosaminidase D [Carbohydrate transport and metabolism] | ||
| pfam03644 | Glyco_hydro_85 | 2.0e-129 | 73 | 357 | 304 | + Glycosyl hydrolase family 85. Family of endo-beta-N-acetylglucosaminidases. These enzymes work on a broad spectrum of substrates. | ||
| cd06547 | GH85_ENGase | 5.0e-144 | 68 | 377 | 330 | + Endo-beta-N-acetylglucosaminidase (ENGase) hydrolyzes the N-N'-diacetylchitobiosyl core of N-glycosylproteins. The beta-1,4-glycosyl bond located between two N-acetylglucosamine residues is hydrolyzed such that N-acetylglucosamine 1 remains with the protein and N-acetylglucosamine 2 forms the reducing end of the released glycan. ENGase is a key enzyme in the processing of free oligosaccharides in the cytosol of eukaryotes. Oligosaccharides formed in the lumen of the endoplasmic reticulum are transported into the cytosol where they are catabolized by cytosolic ENGases and other enzymes, possibly to maximize the reutilization of the component sugars. ENGases have an eight-stranded alpha/beta barrel topology and are classified as a family 85 glycosyl hydrolase (GH85) domain. The GH85 ENGases are sequence-similar to the family 18 glycosyl hydrolases, also known as GH18 chitinases. An ENGase-like protein is also found in bacteria and is included in this alignment model. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005737 | cytoplasm |
| GO:0033925 | mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3fhq_F | 6e-30 | 96 | 453 | 83 | 464 | A Chain A, Structure Of Endo-Beta-N-Acetylglucosaminidase A |
| PDB | 3fhq_D | 6e-30 | 96 | 453 | 83 | 464 | A Chain A, Structure Of Endo-Beta-N-Acetylglucosaminidase A |
| PDB | 3fhq_B | 6e-30 | 96 | 453 | 83 | 464 | A Chain A, Structure Of Endo-Beta-N-Acetylglucosaminidase A |
| PDB | 3fhq_A | 6e-30 | 96 | 453 | 83 | 464 | A Chain A, Structure Of Endo-Beta-N-Acetylglucosaminidase A |
| PDB | 3fha_D | 6e-30 | 96 | 453 | 83 | 464 | A Chain A, Structure Of Endo-Beta-N-Acetylglucosaminidase A |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |