

| Basic Information | |
|---|---|
| Species | Arabidopsis lyrata |
| Cazyme ID | 883240 |
| Family | CBM57 |
| Protein Properties | Length: 2003 Molecular Weight: 220915 Isoelectric Point: 7.4697 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM57 | 1367 | 1536 | 7.8e-24 |
| SINCGGPEIRSVTGALFEREDEDLGPASFVVSAGQRWGASSVGLFAGSSNNIYIATSQSQFVNTLDSELFQSARLSASSLRYYGLGLENGGYTVTLQFAE IQILGSTSNTWRGLGRRRFDIYVQGRLVEKDFDVRRTAGDSTVRAVQREYKANVSENHLEVHLFWAGKGT | |||
| CBM57 | 424 | 594 | 2e-25 |
| VNCGGPDIRSSSGALYEKDEGALGPATFFVSKTQRWAVSNVGLFTGSNSNQYIFVSPTRFANTSDSELFQSARLSASSLRYYGLGLENGGYSVTVQFAEI QIQGSNTWKSLGRRVFDIYVQGKLVEKDFDMHRTANGSSIRVIQRVYKANVSENYLEIHLFWAGKGTCCIP | |||
| Full Sequence |
|---|
| Protein Sequence Length: 2003 Download |
| MPRLILSFVL WFVLMSGLFH VVRPQNRTRA TTDPDEARAL NNIFRTWKIT ATKAWNISGE 60 LCSGAAIDDS VSIDNLAFNP LIKCDCSFVD STICRIVALR ARGMDVAGPI PEDLWTLVYI 120 SNLNLNQNFL TGPLSPGIGN LNRMQWMTFG ANALSGPVPK EIGLLTDLRS LAIDMNNFSG 180 SLPLEIGNCT RLVKMYIGSS GLSGEIPSSF ANFVNLEEAW INDIQLTGQI PDFIGNWTKL 240 TTLRILGTNL SGPIPSTFGN LISLTELRLG EISNINSSLQ FIREMKSISV LVLRNNNLTG 300 TIPSNIGDYL WLRQLDLSFN KLTGQIPAPL FNSRQLTHLF LGNNKLNGSL PTQKSPSLSN 360 IDVSYNDLAG DLPSWVRLPN LQLNLIANHF TVGGSNRRAF RGLDCLQKNF RCNRGKGVYF 420 NFFVNCGGPD IRSSSGALYE KDEGALGPAT FFVSKTQRWA VSNVGLFTGS NSNQYIFVSP 480 TRFANTSDSE LFQSARLSAS SLRYYGLGLE NGGYSVTVQF AEIQIQGSNT WKSLGRRVFD 540 IYVQGKLVEK DFDMHRTANG SSIRVIQRVY KANVSENYLE IHLFWAGKGT CCIPAQGTYG 600 PLVSAISATP DFIPTVKNKL PSKSKKKIGI IVGAIVGAGM LSILVIAIIL FIRRKRKRAA 660 DEEVLNSLHI RPYTFSYSEL RTATQDFDPS NKLGEGGFGP VFKGKLNDGR EIAVKQLSVA 720 SRQGKGQFVA EIATISAVQH RNLVKLYGCC IEGNQRMLVY EYLSNNSLDQ ALFEEKSLQL 780 GWSDRFEICL GVAKGLAYMH EESNPRIVHR DVKASNILLD SDLVPKLSDF GLAKLYDDKK 840 THISTRVAGT IGYLSPEYVM LGHLTEKTDV FAFGIVALEV VSGRPNSSPE LDDDKQYLLE 900 WAWSLHQEKR DLELVDPDLT EFDKEEVKRV IGVAFLCTQT DHAIRPTMSR VVGMLTGDVE 960 VTEANAKPGY VSERTFENAM SFMSGSTSSS WILPETPKDS SKSQDEEHGR RHMLKLRRYL 1020 CLLLTVWFLC NSGSVYVVRA QNRTVATTHP DEARALNSIF AAWRIRAPRE WNISGELCSG 1080 AAIDASVQDS NPAYNPLIKC DCSFENSTIC RITNIKVYAM EVVGPIPQQL WTLEYLTNLN 1140 LGQNVLTGSL PPAIGNLTRM QWMTFGINAL SGPVPKEIGL LTNLKLLSIS SNNFSGSIPD 1200 EIGRCTKLQQ IYIDSSGLSG RIPVSFANLV ELEQAWIADM ELTGQIPDFI GDWTNLTTLR 1260 ILGTGLSGPI PASFSNLTSL TELFLGNNTL NGSLPTQKRQ SLSNIDVSYN DLSGSLPSWV 1320 SLPNLNLNLV ANNFTLEGLD NRVLSGLNCL QKNFPCNRGK GIYSDFSINC GGPEIRSVTG 1380 ALFEREDEDL GPASFVVSAG QRWGASSVGL FAGSSNNIYI ATSQSQFVNT LDSELFQSAR 1440 LSASSLRYYG LGLENGGYTV TLQFAEIQIL GSTSNTWRGL GRRRFDIYVQ GRLVEKDFDV 1500 RRTAGDSTVR AVQREYKANV SENHLEVHLF WAGKGTCCIP IQGAYGPLIS AVGATPDFTP 1560 TVGNRPPSKG KSMTGTIVGV IVGVGLLSIF AGVVIFIIRK RRKRYTDDEE ILSMDVKPYT 1620 FTYSELKSAT QDFDPSNKLG EGGFGPVYKG KLNDGREIAV KLLSVGSRQG KGQFVAEIVA 1680 ISAVQHRNLV KLYGCCYEGD HRLLVYEYLP NGSLDQALFG THRNMIIDLC FCQPKSTHYV 1740 LVVGLNVAGE KTLHLDWSTR YEICLGVARG LVYLHEEARL RIVHRDVKAS NILLDSKLVP 1800 KVSDFGLAKL YDDKKTHIST RVAGTIGYLA PEYAMRGHLT EKTDVYAFGV VALELVSGRP 1860 NSDENLEDEK RYLLEWAWNL HEKSREVELI DHELTDFNTE EAKRMIGIAL LCTQTSHALR 1920 PPMSRVVAML SGDVEVSDVT SKPGYLTDWR FDDTTGSSIS GFRIKTTEAS ESFMSFVAPG 1980 SEISPRNNDS KPMLGAQMNE GR* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
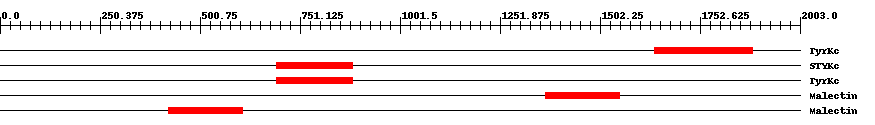 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| smart00219 | TyrKc | 6.0e-47 | 1638 | 1885 | 275 | + Tyrosine kinase, catalytic domain. Phosphotransferases. Tyrosine-specific kinase subfamily. | ||
| smart00221 | STYKc | 6.0e-48 | 692 | 882 | 197 | + Protein kinase; unclassified specificity. Phosphotransferases. The specificity of this class of kinases can not be predicted. Possible dual-specificity Ser/Thr/Tyr kinase. | ||
| smart00219 | TyrKc | 4.0e-48 | 692 | 882 | 197 | + Tyrosine kinase, catalytic domain. Phosphotransferases. Tyrosine-specific kinase subfamily. | ||
| pfam11721 | Malectin | 5.0e-52 | 1366 | 1552 | 189 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| pfam11721 | Malectin | 9.0e-57 | 421 | 606 | 187 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004672 | protein kinase activity |
| GO:0005515 | protein binding |
| GO:0005524 | ATP binding |
| GO:0006468 | protein phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAG50909.1 | 0 | 4 | 981 | 1024 | 2016 | AC069159_10 receptor protein kinase, putative [Arabidopsis thaliana] |
| GenBank | AAG50909.1 | 0 | 15 | 2002 | 1 | 2062 | AC069159_10 receptor protein kinase, putative [Arabidopsis thaliana] |
| EMBL | CBI20016.1 | 0 | 13 | 984 | 1169 | 2155 | unnamed protein product [Vitis vinifera] |
| EMBL | CBI20016.1 | 0 | 30 | 1955 | 104 | 2151 | unnamed protein product [Vitis vinifera] |
| RefSeq | NP_564710.1 | 0 | 1 | 1012 | 1 | 1012 | leucine-rich repeat family protein / protein kinase family protein [Arabidopsis thaliana] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3ulz_A | 0 | 675 | 958 | 20 | 309 | A Chain A, Lip5(mit)2 |
| PDB | 3ulz_A | 0 | 1621 | 1933 | 20 | 309 | A Chain A, Lip5(mit)2 |
| PDB | 3uim_A | 0 | 675 | 958 | 20 | 309 | A Chain A, Structural Basis For The Impact Of Phosphorylation On Plant Receptor- Like Kinase Bak1 Activation |
| PDB | 3uim_A | 0 | 1621 | 1933 | 20 | 309 | A Chain A, Structural Basis For The Impact Of Phosphorylation On Plant Receptor- Like Kinase Bak1 Activation |
| PDB | 3tl8_H | 0 | 675 | 958 | 28 | 317 | B Chain B, The Avrptob-Bak1 Complex Reveals Two Structurally Similar Kinaseinteracting Domains In A Single Type Iii Effector |