

| Basic Information | |
|---|---|
| Species | Eucalyptus grandis |
| Cazyme ID | Eucgr.B00862.1 |
| Family | AA1 |
| Protein Properties | Length: 588 Molecular Weight: 65311.4 Isoelectric Point: 6.6416 |
| Chromosome | Chromosome/Scaffold: 2 Start: 10511217 End: 10513513 |
| Description | Cupredoxin superfamily protein |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 99 | 361 | 1e-22 |
| ANATVPGPTIEAIQNVPTYVTWQNFLPQSHILPWDPTIPTAIPKNGGVPTVVHLHGGISPPQSDGHPYAWFTANFAETGPTWTQQTYTYPNAQHAGNLWY HDHAVGLTRVNLLAGLIAPYIIRNLPLDARLNLPLGPEFDRVLMIFDRSFNTDGSLYMNSTGVNPTIHPQWQPEYFGDAIIVNGKAWPYLQVQRRRYRFR IINTSNARYYRLSLTNNLTFTQIGSDGSYLPKPVGTQTVLLGPSEGADVVVDFSMTAANETIL | |||
| Full Sequence |
|---|
| Protein Sequence Length: 588 Download |
| MNTGLLSSWI LLMILVGWAL TEAQTPPPPP VTDTTLFQEA ASLQMYVDEL PQIPKLFGYT 60 MSNGVPRPAN LTIGMYKTTW KFHRDLPATT VFAYGTSAAN ATVPGPTIEA IQNVPTYVTW 120 QNFLPQSHIL PWDPTIPTAI PKNGGVPTVV HLHGGISPPQ SDGHPYAWFT ANFAETGPTW 180 TQQTYTYPNA QHAGNLWYHD HAVGLTRVNL LAGLIAPYII RNLPLDARLN LPLGPEFDRV 240 LMIFDRSFNT DGSLYMNSTG VNPTIHPQWQ PEYFGDAIIV NGKAWPYLQV QRRRYRFRII 300 NTSNARYYRL SLTNNLTFTQ IGSDGSYLPK PVGTQTVLLG PSEGADVVVD FSMTAANETI 360 LTNDAPYAYP SGNPVDQLNS KVMKFIIQPR APIQTDNSHV PPNLKTYPVA SNAFGMAKTT 420 RYITLYEYLT PAGTSTHLYI NGLRFEDPVT ETPRVGTTEI WEVINLTGDN HPFHMHLAAF 480 QAVKIQQLVD LAGFTSCMTQ LNDAIKCNIT GHATGPLVDV PLTERTWKNV VKMAPGYKTT 540 LVVQFFVVDD AYSTYPFDAT AEPGYVYHCH ILDHEDNAMI RPMKLVH* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
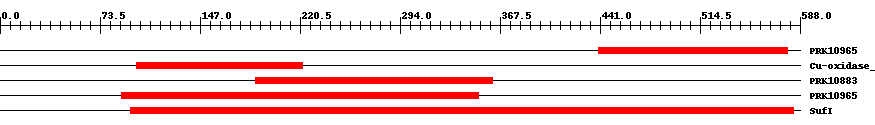 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK10965 | PRK10965 | 7.0e-5 | 440 | 579 | 144 | + multicopper oxidase; Provisional | ||
| pfam07732 | Cu-oxidase_3 | 2.0e-7 | 100 | 222 | 125 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| PRK10883 | PRK10883 | 5.0e-15 | 188 | 362 | 182 | + FtsI repressor; Provisional | ||
| PRK10965 | PRK10965 | 6.0e-16 | 89 | 352 | 276 | + multicopper oxidase; Provisional | ||
| COG2132 | SufI | 1.0e-44 | 96 | 583 | 499 | + Putative multicopper oxidases [Secondary metabolites biosynthesis, transport, and catabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| DDBJ | BAB21194.1 | 0 | 31 | 586 | 34 | 587 | putative spore coat protein-like protein [Oryza sativa Japonica Group] |
| GenBank | EAZ10376.1 | 0 | 28 | 571 | 27 | 566 | hypothetical protein OsJ_00212 [Oryza sativa Japonica Group] |
| RefSeq | NP_001041895.1 | 0 | 28 | 586 | 27 | 581 | Os01g0126100 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002455264.1 | 0 | 30 | 586 | 33 | 587 | hypothetical protein SORBIDRAFT_03g007440 [Sorghum bicolor] |
| RefSeq | XP_002457447.1 | 0 | 29 | 586 | 26 | 588 | hypothetical protein SORBIDRAFT_03g007480 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3zdw_A | 0 | 42 | 585 | 2 | 508 | A Chain A, Rice Bglu1 Beta-Glucosidase, A Plant ExoglucanaseBETA-Glucosidase |
| PDB | 2x88_A | 0 | 42 | 585 | 2 | 508 | A Chain A, Rice Bglu1 Beta-Glucosidase, A Plant ExoglucanaseBETA-Glucosidase |
| PDB | 2x87_A | 0 | 42 | 585 | 2 | 508 | A Chain A, Crystal Structure Of The Reconstituted Cota |
| PDB | 2bhf_A | 0 | 42 | 585 | 2 | 508 | A Chain A, Crystal Structure Of The Reconstituted Cota |
| PDB | 1w8e_A | 0 | 42 | 585 | 2 | 508 | A Chain A, Crystal Structure Of The Reconstituted Cota |