

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s110_116V6.1 |
| Family | GT1 |
| Protein Properties | Length: 849 Molecular Weight: 93092.9 Isoelectric Point: 7.6715 |
| Chromosome | Chromosome/Scaffold: 110 Start: 1058361 End: 1064217 |
| Description | UDP-Glycosyltransferase superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 395 | 804 | 0 |
| IVGTRGDVQPFVAIGKQLQEYGHRVRLATHTNFRDFVKKEGLEFYPLGGDPKVLAEYMVKNKGFLPSGPSEVSVQRKQIKSIVYSLLDACIKPDKESGVP FRPHAIIANPPAYGHVHVAEYLQIPLHIFFTMPWTSTREFPHPLSRIKQPAGNRMSYQVVDSLIWLGIRGIINSFRKKQLKLRPITYLSGSQGSIADLPT GYIWSPHLVPKPKDWGPLVDVVGFCFLNLAQNYKPPDDLVKWLDAGPPPIYIGFGSLPVADPEGMTKIIIEALEKTAQRGIIGRGWGGIGNLPEVPDNVY LLSDCPHDWLFPRCAAVVHHGGAGTTAAGLRAACPTTIVPFFGDQPFWGERVQQKGVGPAHIPVKHFDLEKLVSAIEFMLDPTVKQAALTLANSMKGEDG IKGAVNVFHK | |||
| Full Sequence |
|---|
| Protein Sequence Length: 849 Download |
| MTLSGERDVG VSSYVPPGTS STASKPLPNS PQVPLQTTFP AQVSSKALPN LAADAGSGNN 60 NESSGIISAL LSKDKNESAL SQVDLSDIAI ETWPTRGRPK ENVLNEVPHV NEILNPTGTR 120 SKDDLFNDSD GNMAYGNGGD KGDSEGGKIS LLGPKSKDST KVFATHSDSH LPGPIGDTQH 180 HRDPQPFPLH GPQYDLQPGS EQKTPIEAGR PLQATQSLPV WMPSTVTSTD APESIAEEVE 240 ALVTPFPTPQ AIEEVDQSSL GGTPAKDQMP RTTSMPEADF QMSERIRQDH LTQSNILKRR 300 INKNRDNANV RVAARSKLLD RKKKKLLRQF AMIQTDGGVE FDLRQSGQLA RDLFGDHVMD 360 EEESDHGIPA ANRIEEGNEE NDQSPVPPLK IVMLIVGTRG DVQPFVAIGK QLQEYGHRVR 420 LATHTNFRDF VKKEGLEFYP LGGDPKVLAE YMVKNKGFLP SGPSEVSVQR KQIKSIVYSL 480 LDACIKPDKE SGVPFRPHAI IANPPAYGHV HVAEYLQIPL HIFFTMPWTS TREFPHPLSR 540 IKQPAGNRMS YQVVDSLIWL GIRGIINSFR KKQLKLRPIT YLSGSQGSIA DLPTGYIWSP 600 HLVPKPKDWG PLVDVVGFCF LNLAQNYKPP DDLVKWLDAG PPPIYIGFGS LPVADPEGMT 660 KIIIEALEKT AQRGIIGRGW GGIGNLPEVP DNVYLLSDCP HDWLFPRCAA VVHHGGAGTT 720 AAGLRAACPT TIVPFFGDQP FWGERVQQKG VGPAHIPVKH FDLEKLVSAI EFMLDPTVKQ 780 AALTLANSMK GEDGIKGAVN VFHKHIHKHV PNILKLTPLE DLHRHHGIFF RIKRKCKSMM 840 HRSSHNSS* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG1819 | COG1819 | 1.0e-28 | 390 | 804 | 429 | + Glycosyl transferases, related to UDP-glucuronosyltransferase [Carbohydrate transport and metabolism / Signal transduction mechanisms] | ||
| pfam03033 | Glyco_transf_28 | 1.0e-31 | 391 | 535 | 145 | + Glycosyltransferase family 28 N-terminal domain. The glycosyltransferase family 28 includes monogalactosyldiacylglycerol synthase (EC 2.4.1.46) and UDP-N-acetylglucosamine transferase (EC 2.4.1.-). This N-terminal domain contains the acceptor binding site and likely membrane association site. This family also contains a large number of proteins that probably have quite distinct activities. | ||
| cd03784 | GT1_Gtf_like | 5.0e-109 | 390 | 805 | 420 | + This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005975 | carbohydrate metabolic process |
| GO:0008152 | metabolic process |
| GO:0016758 | transferase activity, transferring hexosyl groups |
| GO:0030259 | lipid glycosylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI37267.1 | 0 | 269 | 806 | 58 | 589 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_001769124.1 | 0 | 325 | 848 | 1 | 524 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001773142.1 | 0 | 359 | 844 | 1 | 488 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001778784.1 | 0 | 317 | 814 | 12 | 512 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001780556.1 | 0 | 325 | 840 | 1 | 516 | predicted protein [Physcomitrella patens subsp. patens] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3h4t_A | 2e-30 | 389 | 809 | 1 | 384 | A Chain A, Crystal Structure Of Mutation Of An Acylptide HydrolaseESTERASE FROM AEROPYRUM PERNIX K1 |
| PDB | 3h4i_A | 2e-30 | 389 | 809 | 1 | 384 | A Chain A, Chimeric Glycosyltransferase For The Generation Of Novel Natural Products |
| PDB | 1iir_A | 1e-26 | 632 | 816 | 229 | 408 | A Chain A, Crystal Structure Of Udp-Glucosyltransferase Gtfb |
| PDB | 1rrv_B | 9e-23 | 389 | 787 | 1 | 381 | A Chain A, X-Ray Crystal Structure Of Tdp-Vancosaminyltransferase Gtfd As A Complex With Tdp And The Natural Substrate, Desvancosaminyl Vancomycin. |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
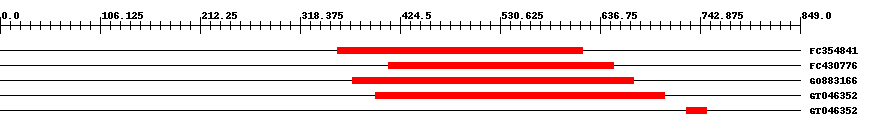 | ||||
| Hit | Length | Start | End | EValue |
| FC354841 | 261 | 358 | 618 | 0 |
| FC430776 | 240 | 412 | 651 | 0 |
| GO883166 | 299 | 374 | 672 | 0 |
| GT046352 | 308 | 398 | 705 | 0 |
| GT046352 | 22 | 729 | 750 | 0.28 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |