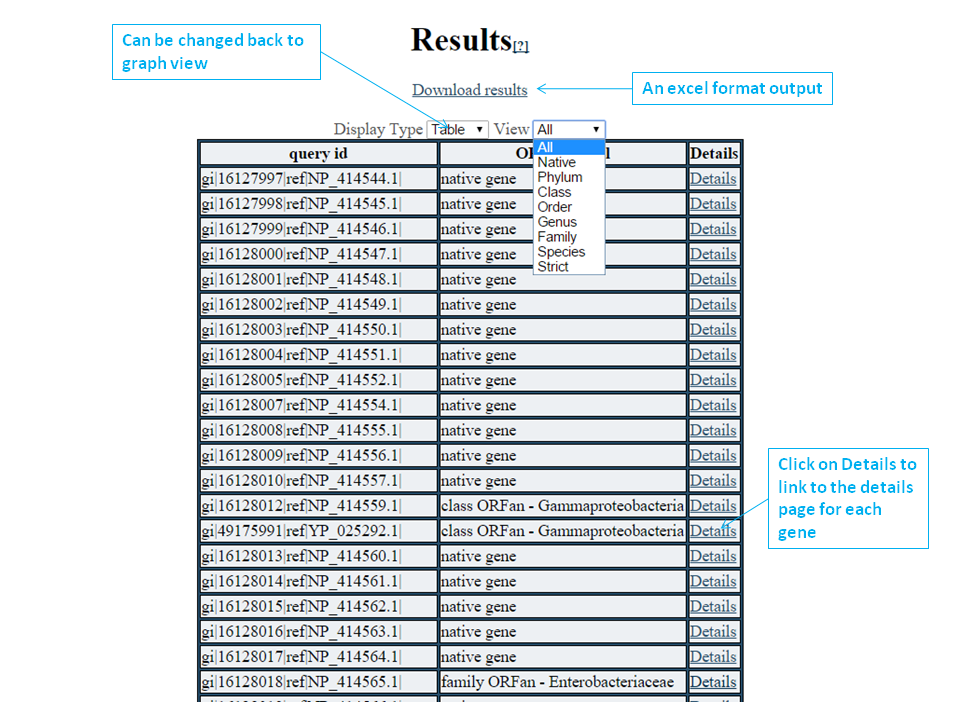
What is ORFanFinder?
ORFanFinder is a tool that determines the phylogenetic level at which a gene is unique. In other words, given a genome, ORFanFinder can classify its gene content into groups of different ages with a special focus on ORFans. Read the Manual for more method details. We provide this web server for users to get this information graphically, as well as a downloadable command line version.What are ORFans?
Definition: Orphan genes are new protein-coding genes that are restricted to taxonomically closely related genomes. A synonymous term ORFan (orphan Open Reading Frames) was introduced by (Fischer and Eisenberg, 1999) and is more commonly used in the microbial literature.Orphan genes are new inventions of a genome: Orphan genes are also known as new genes or ORFans that enable the organism to adapt to its specific living environment (Khalturin et al., 2009; Chen et al., 2013). In the past decade, elucidating the origin, evolution and function of orphan genes has become a highly interesting research topic in the evolutionary genomics field (Tautz and Domazet-Loso, 2011; Carvunis et al., 2012; Long et al., 2013).
Relevance to pathogenesis: In the microbial genomics field, recent pan-genome analyses of numerous bacterial pathogens and their closely related laboratory strains have suggested that each genome of a same species may have a significant fraction of its gene content to be unique (Medini et al., 2005; Medini et al., 2008; Bentley, 2009). Many of the unique genes are lineage-specific ORFans that are shown to reside in pathogenic islands or prophages (phages incorporated into the host genomes) and thus may contribute to pathogenesis (Raskin et al., 2006; Pallen and Wren, 2007).
ORFans of different ages: From the evolutionary genomics perspective any genome is a gene pool that has been dynamically changing throughout its evolution. As a result, some genes of a genome may have emerged very recently only in the studied genome (genome-specific ORFans), while some others may have firstly appeared in the most recent common ancestor of an entire genus of species (genus-specific ORFans). Hence to understand how ORFans have contributed to the niche-specific adaptation and pathogenesis, computer tools are desired to automatically identify, classify, annotate and present ORFans of different ages, which will assist further evolutionary and functional analyses.
News
2016/03: ORFanFinder program and web server paper is accepted for publication at Bioinformatics2015/03: ORFanFinder got funded by NIH (grant #: 1R15GM114706)
2014/06: Devleoped ORFanFinder server v1.0
2014/01: Developed a new taxonomy-based algorithm for determining the age of ORFans
2008/01: Published the first genome-wide investigation of ORFans in viral genomes (Yin and Fischer, 2008)
2006/08: Published an algorithm for identifying ORFans in prokaryotic genomes (Yin and Fischer, 2006)
About us
Alex Ekstrom: Research Assistant of Computer ScienceDr. Yanbin Yin: Assistant Professor of Bioinformatics
Similar web resources
- The ORFanage: an ORFan database (Siew et al., 2004) (obsolete website)- OrphanMine: Taxonomically Restricted Genes (TRGs) database in Bacterial and Archaeal genomes (Wilson et al., 2007) (obsolete website)
- ProteinHistorian: Protein Age Estimation and Enrichment Analysis (Capra et al., 2012)
- GenTree database: Human lineage-specific genes (website)