

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma09g23706.1 |
| Family | GT1 |
| Protein Properties | Length: 332 Molecular Weight: 36908.1 Isoelectric Point: 8.2759 |
| Chromosome | Chromosome/Scaffold: 09 Start: 29338260 End: 29340652 |
| Description | UDP-glucosyl transferase 88A1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT1 | 207 | 299 | 2e-25 |
| APVVLLCYGSMGRFSRAQLKEIALGLEKSEQRFLWVAPQVQILSHDSVGGFVTHCGWNLVLEAMCEGVPMVVWPLYTEQKMNRVILVKEIKVA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 332 Download |
| MKDTIVLYPN LGRGHLVSMV ELGKLILTQY PSLSITILIL TPPITPSTTT IACDSNARYI 60 TTVTATTPAI TFHHVPFNFN TPSLPLHILS LEFTRHSTQN ITVALQTLAK ASNLKALVMD 120 FMNFNDPKAL TKNLNNNVPI YFYYTSCAST LSTPSFYQAL LQLPENMEDG VGIITNTFEG 180 IEEKPIRTLS KDVTIPPLFC VRPMISAPVV LLCYGSMGRF SRAQLKEIAL GLEKSEQRFL 240 WVAPQVQILS HDSVGGFVTH CGWNLVLEAM CEGVPMVVWP LYTEQKMNRV ILVKEIKVAL 300 AVNENKEGFV SVTELVKGCQ WWRGLSTQSR R* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
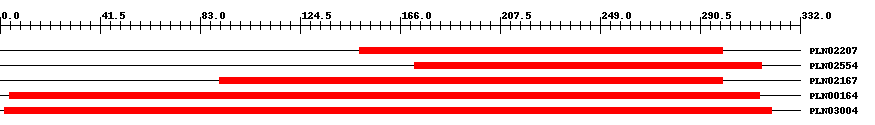 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02207 | PLN02207 | 2.0e-29 | 149 | 300 | 207 | + UDP-glycosyltransferase | ||
| PLN02554 | PLN02554 | 2.0e-31 | 172 | 316 | 213 | + UDP-glycosyltransferase family protein | ||
| PLN02167 | PLN02167 | 1.0e-31 | 91 | 300 | 316 | + UDP-glycosyltransferase family protein | ||
| PLN00164 | PLN00164 | 1.0e-41 | 4 | 315 | 434 | + glucosyltransferase; Provisional | ||
| PLN03004 | PLN03004 | 2.0e-60 | 2 | 320 | 427 | + UDP-glycosyltransferase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0008152 | metabolic process |
| GO:0016758 | transferase activity, transferring hexosyl groups |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABI94021.1 | 0 | 1 | 315 | 1 | 413 | (iso)flavonoid glycosyltransferase [Medicago truncatula] |
| GenBank | ABI94022.1 | 0 | 1 | 315 | 1 | 416 | (iso)flavonoid glycosyltransferase [Medicago truncatula] |
| GenBank | ACJ72159.1 | 0 | 1 | 315 | 3 | 421 | UGT2 [Pueraria montana var. lobata] |
| DDBJ | BAB86923.1 | 0 | 1 | 315 | 1 | 415 | glucosyltransferase-5 [Vigna angularis] |
| DDBJ | BAF64416.1 | 0 | 1 | 315 | 1 | 423 | UDP-glucose:isoflavone 7-O-glucosyltransferase [Glycine max] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2acw_B | 5e-36 | 5 | 320 | 12 | 420 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2acw_A | 5e-36 | 5 | 320 | 12 | 420 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2acv_B | 5e-36 | 5 | 320 | 12 | 420 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2acv_A | 5e-36 | 5 | 320 | 12 | 420 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| PDB | 2vg8_A | 1e-30 | 5 | 318 | 9 | 425 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| rose anthocyanin biosynthesis | RXN-8005 | EC-2.4.1 | UDP-D-glucose:cyanidin 5-O-β-D-glucosyltransferase |
| rose anthocyanin biosynthesis | RXN-8006 | EC-2.4.1 | cyanidin 5,3-O-glycosyltransferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| FE686907 | 216 | 158 | 315 | 0 |
| HO798145 | 281 | 132 | 314 | 0 |
| BI973336 | 183 | 192 | 315 | 0 |
| GR853891 | 220 | 157 | 315 | 0 |
| BI973336 | 19 | 314 | 332 | 6.8 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |