

| Basic Information | |
|---|---|
| Species | Ricinus communis |
| Cazyme ID | 29222.m000396 |
| Family | GT24 |
| Protein Properties | Length: 1512 Molecular Weight: 171304 Isoelectric Point: 6.0507 |
| Chromosome | Chromosome/Scaffold: 29222 Start: 52376 End: 79553 |
| Description | UDP-glucose:glycoprotein glucosyltransferases;transferases, transferring hexosyl groups;transferases, transferring glycosyl groups |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT24 | 1302 | 1499 | 0 |
| INIFSIASGHLYERFLKIMILSVLKNTQRPVKFWFIKNYLSPQFKDVIPCMAQEYGFEYELITYKWPSWLHKQKEKQRIIWAYKILFLDVIFPLSLEKVI FVDADQVVRADMGELYDMDIKGRPLAYTPFCDNNKDMDGYRFWRQGFWKEHLRGRPYHISALYVVDLVKFRETAAGDNLRVFYETLSKDPNSLANLDQ | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1512 Download |
| MGTRFRSGFC VLIIVLCVSF CGLFSVHGEN RRPKNVQVAI RAKWEGTPVL LEAGELLSKE 60 RRDLYWAFIE VWLQAENDEP DSYTAKNCLR RIIKHGNSLL SDPLASLFEF SLILRSASPR 120 LVLYRQLAEE SLSSFPFLDD SISDNARKCC WVDTGGALFF DVAEVLLWLK NPAKLAGDPF 180 QQPELFDFDH VHFDSQTGSP VAILYGALGT DCFREFHTTL AQAAKEGKVK YIVRPVLPSG 240 CEAKVSHCGA IGSEESLNLG GYGVELALKN MEYKAMDDSA IKKGVTLEDP RTEDLTQEVR 300 GFIFSKLLER KPELTSEIMA FRDYLLSSTI SDTLDVWELK DLGHQTAQRI VHASDPLQSM 360 QEINQNFPSI VSYLSRMKLN DSIKDEITAN QRMIPPGKSL MALNGALINV EDIDLYLLID 420 MVQQELLLAD QFSKMKVPHS TIRKLLSTMS PPESNMFRVD FRSTHVHYLN NLEEDAMYKQ 480 WRSNINEILM PVFPGQLRYI RKNLFHAVYV LDPATSCGLE ASDFFSPFTN NYPLNPFIKK 540 IEVSGGDLHL SSIEDNSQTE EDLSSLIIRL FIYIKENYGM KTAFQFLSNV NRLRVESAES 600 VDDAPEMHNV EGGFVEAILS KVKSPPQDIL LKLEKEKEFK ELSQESSVAV FKLGLYKLQC 660 CLLMNGLVSD SREEALMIAM NDELPRIQEQ VYYGHINSRT DILDKFLSES SISRYNPQII 720 AEGKGKPRFI SLSSSVLDGE SVIHDISYLH SSETVDDLKP VTQLLVVDLT SLRGIKLLHE 780 GILYLIRGSK VARLGVLFSA SRDADLPSLL IAKVFEITVS SYSHKKNVLH FLEQLCSFYE 840 QSGVHASSLT DESSQAFIEK VYELADANEL SRKAYKSALT EFSIDAMKRH LDKVAKLLYR 900 QLGLEAGVSA IITNGRVTIL NDVGTFLSHD LNLLESVEFK QRIKHIVEII EEVHWQDIDP 960 DMLTSKFVSD IVMTVSSAMA LRDRSSESAR FEILNADYSA VILENENSSV HIDAVVDPLS 1020 PVGQHVASLL KVLRQYIQPS MRIVLNPMSS LVDLPLKNFY RYVVPTMDDF SSTDHTINGP 1080 KAFFANMPLS KTLTMNLDVP EPWLVEPVIA VHDLDNILLE NLGDTRTLQA IFELEALVLT 1140 GHCSEKDQEP PRGLQLILGT KGAPHLVDTI VMANLGYWQM KVSPGVWYLQ LAPGRSSELY 1200 VLKEDGAESL DKLLSKRITI NDFRGKVVHL EVAKKKGMEH EKLLVPSDDD NHMHRNKKGT 1260 HNSWNSNLLK WASGFIGGNG LAKKNENVLV EHAKGSRRGK PINIFSIASG HLYERFLKIM 1320 ILSVLKNTQR PVKFWFIKNY LSPQFKDVIP CMAQEYGFEY ELITYKWPSW LHKQKEKQRI 1380 IWAYKILFLD VIFPLSLEKV IFVDADQVVR ADMGELYDMD IKGRPLAYTP FCDNNKDMDG 1440 YRFWRQGFWK EHLRGRPYHI SALYVVDLVK FRETAAGDNL RVFYETLSKD PNSLANLDQL 1500 SMRMLTGIQG EN |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
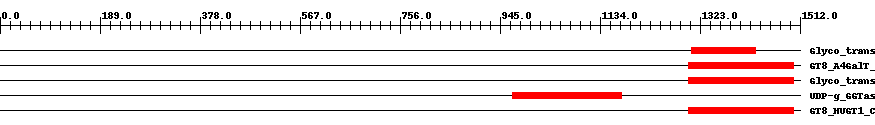 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam01501 | Glyco_transf_8 | 0.0006 | 1306 | 1427 | 128 | + Glycosyl transferase family 8. This family includes enzymes that transfer sugar residues to donor molecules. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. This family includes Lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, and glycogenin glucosyltransferase. | ||
| cd04194 | GT8_A4GalT_like | 3.0e-11 | 1302 | 1499 | 206 | + A4GalT_like proteins catalyze the addition of galactose or glucose residues to the lipooligosaccharide (LOS) or lipopolysaccharide (LPS) of the bacterial cell surface. The members of this family of glycosyltransferases catalyze the addition of galactose or glucose residues to the lipooligosaccharide (LOS) or lipopolysaccharide (LPS) of the bacterial cell surface. The enzymes exhibit broad substrate specificities. The known functions found in this family include: Alpha-1,4-galactosyltransferase, LOS-alpha-1,3-D-galactosyltransferase, UDP-glucose:(galactosyl) LPS alpha1,2-glucosyltransferase, UDP-galactose: (glucosyl) LPS alpha1,2-galactosyltransferase, and UDP-glucose:(glucosyl) LPS alpha1,2-glucosyltransferase. Alpha-1,4-galactosyltransferase from N. meningitidis adds an alpha-galactose from UDP-Gal (the donor) to a terminal lactose (the acceptor) of the LOS structure of outer membrane. LOSs are virulence factors that enable the organism to evade the immune system of host cells. In E. coli, the three alpha-1,2-glycosyltransferases, that are involved in the synthesis of the outer core region of the LPS, are all members of this family. The three enzymes share 40 % of sequence identity, but have different sugar donor or acceptor specificities, representing the structural diversity of LPS. | ||
| cd00505 | Glyco_transf_8 | 3.0e-42 | 1302 | 1499 | 199 | + Members of glycosyltransferase family 8 (GT-8) are involved in lipopolysaccharide biosynthesis and glycogen synthesis. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. GT-8 comprises enzymes with a number of known activities: lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, glycogenin glucosyltransferase, and N-acetylglucosaminyltransferase. GT-8 enzymes contains a conserved DXD motif which is essential in the coordination of a catalytic divalent cation, most commonly Mn2+. | ||
| pfam06427 | UDP-g_GGTase | 4.0e-80 | 969 | 1174 | 212 | + UDP-glucose:Glycoprotein Glucosyltransferase. The N-terminal region of this group of proteins is required for correct folding of the ER UDP-Glc: glucosyltransferase. | ||
| cd06432 | GT8_HUGT1_C_like | 7.0e-134 | 1302 | 1499 | 198 | + The C-terminal domain of HUGT1-like is highly homologous to the GT 8 family. C-terminal domain of glycoprotein glucosyltransferase (UGT). UGT is a large glycoprotein whose C-terminus contains the catalytic activity. This catalytic C-terminal domain is highly homologous to Glycosyltransferase Family 8 (GT 8) and contains the DXD motif that coordinates donor sugar binding, characteristic for Family 8 glycosyltransferases. GT 8 proteins are retaining enzymes based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. The non-catalytic N-terminal portion of the human UTG1 (HUGT1) has been shown to monitor the protein folding status and activate its glucosyltransferase activity. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity |
| GO:0006486 | protein glycosylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAG51883.1 | 0 | 1 | 1499 | 1 | 1541 | AC016162_4 putative UDP-glucose:glycoprotein glucosyltransferase; 101200-91134 [Arabidopsis thaliana] |
| EMBL | CBI23772.1 | 0 | 1 | 1290 | 1 | 1301 | unnamed protein product [Vitis vinifera] |
| RefSeq | NP_177278.3 | 0 | 1 | 1499 | 1 | 1498 | EBS1 (EMS-mutagenized bri1 suppressor 1); UDP-glucose:glycoprotein glucosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups [Arabidopsis thaliana] |
| RefSeq | XP_002268972.1 | 0 | 1 | 1499 | 1 | 1501 | PREDICTED: similar to EBS1 (EMS-MUTAGENIZED BRI1 SUPPRESSOR 1); UDP-glucose:glycoprotein glucosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups [Vitis vinifera] |
| RefSeq | XP_002529534.1 | 0 | 1 | 1512 | 1 | 1512 | UDP-glucose glycoprotein:glucosyltransferase, putative [Ricinus communis] |