

| Basic Information | |
|---|---|
| Species | Ricinus communis |
| Cazyme ID | 30032.m000475 |
| Family | GT41 |
| Protein Properties | Length: 930 Molecular Weight: 103624 Isoelectric Point: 5.7704 |
| Chromosome | Chromosome/Scaffold: 30032 Start: 139573 End: 149527 |
| Description | Tetratricopeptide repeat (TPR)-like superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT41 | 364 | 853 | 0 |
| IKPNFSQSLNNLGVVYTVQGKMDAAASMIEKAIMANPTYAEAYNNLGVLYRDAGNIPMAINAYEQCLKIDPDSRNAGQNRLLAMNYINEGHDEKLFEAHR DWGRRFMRLYPQYTMWDNPKDLDRPLVIGYVSPDYFTHSVSYFIEAPLVYHDYANYKVVVYSAVVKADAKTIRFREKVLKQGGIWRDIYGIDEKKVASMV REDNVDILVELTGHTANNKLGMMACRPAPIQVTWIGYPNTTGLPTIDYRITDSLADPRDTKQKHVEELVRLPDCFLCYTPSPEAGPVCPTPALANGFITF GSFNNLAKITPKVLQVWARILCAVPNSRLVVKCKPFCCDSVRQRFLTMLEELGLESLRVDLLPLILLNHDHMQAYSLMDISLDTFPYAGTTTTCESLYMG VPCVTMAGAIHAHNVGVSLLSKVGLGHLVAQNEDNYVQLALQLASDIPALSNLRMSLRDLMSKSPVCDGSKFTLGLESSYRDMWHRYCKG | |||
| Full Sequence |
|---|
| Protein Sequence Length: 930 Download |
| MAWTEKNNGN GKEGGPIEDN GFLKGTQEPS PSASGSPVAV AAGLKGIEEK DSLSYANILR 60 SRNKFVDALA IYESVLEKDS GNVEAYIGKG ICLQMQNMGR LAFDSFAEAI KLDPQNACAL 120 THCGILYKEE GRLVEAAESY QKALRADPLY KPAAECLSIV LTDLGTSLKL SGNTQEGIQK 180 YYEALKIDPH YAPAYYNLGV VYSEMMQYDT ALNCYEKAAL ERPMYAEAYC NMGVIYKNRG 240 DLESAIACYE RCLAVSPNFE IAKNNMAIAL TDLGTKVKLE GDINQGIAYY KKALYYNWHY 300 ADAMYNLGVA YGEMLKFDNA IVFYELAFHF NPHCAEACNN LGVIYKDRDN LDKAVECYQT 360 ALSIKPNFSQ SLNNLGVVYT VQGKMDAAAS MIEKAIMANP TYAEAYNNLG VLYRDAGNIP 420 MAINAYEQCL KIDPDSRNAG QNRLLAMNYI NEGHDEKLFE AHRDWGRRFM RLYPQYTMWD 480 NPKDLDRPLV IGYVSPDYFT HSVSYFIEAP LVYHDYANYK VVVYSAVVKA DAKTIRFREK 540 VLKQGGIWRD IYGIDEKKVA SMVREDNVDI LVELTGHTAN NKLGMMACRP APIQVTWIGY 600 PNTTGLPTID YRITDSLADP RDTKQKHVEE LVRLPDCFLC YTPSPEAGPV CPTPALANGF 660 ITFGSFNNLA KITPKVLQVW ARILCAVPNS RLVVKCKPFC CDSVRQRFLT MLEELGLESL 720 RVDLLPLILL NHDHMQAYSL MDISLDTFPY AGTTTTCESL YMGVPCVTMA GAIHAHNVGV 780 SLLSKVGLGH LVAQNEDNYV QLALQLASDI PALSNLRMSL RDLMSKSPVC DGSKFTLGLE 840 SSYRDMWHRY CKGDVPSLKR MELLKQQKGS EAVPNENFEP TRNAFPVEGP PESVKLNGYN 900 IVSSSILNRS SEENVSQTQL NHTTNSDKPS |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| TIGR02917 | PEP_TPR_lipo | 6.0e-21 | 51 | 453 | 437 | + putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. | ||
| TIGR02917 | PEP_TPR_lipo | 1.0e-21 | 109 | 448 | 343 | + putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. | ||
| TIGR02917 | PEP_TPR_lipo | 2.0e-23 | 53 | 435 | 417 | + putative PEP-CTERM system TPR-repeat lipoprotein. This protein family occurs in strictly within a subset of Gram-negative bacterial species with the proposed PEP-CTERM/exosortase system, analogous to the LPXTG/sortase system common in Gram-positive bacteria. This protein occurs in a species if and only if a transmembrane histidine kinase (TIGR02916) and a DNA-binding response regulator (TIGR02915) also occur. The present of tetratricopeptide repeats (TPR) suggests protein-protein interaction, possibly for the regulation of PEP-CTERM protein expression, since many PEP-CTERM proteins in these genomes are preceded by a proposed DNA binding site for the response regulator. | ||
| COG3914 | Spy | 6.0e-84 | 318 | 850 | 562 | + Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005515 | protein binding |
| GO:0042802 | identical protein binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAX46402.1 | 0 | 1 | 914 | 1 | 914 | putative SPINDLY protein [Rosa luciae] |
| RefSeq | XP_002281883.1 | 0 | 1 | 914 | 1 | 911 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002308458.1 | 0 | 1 | 930 | 1 | 934 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002323412.1 | 0 | 1 | 903 | 1 | 906 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002530535.1 | 0 | 1 | 930 | 1 | 930 | o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 4gz6_D | 6e-39 | 300 | 639 | 8 | 386 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 4gz6_D | 4e-22 | 661 | 856 | 524 | 715 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 4gz6_D | 5e-19 | 114 | 270 | 6 | 155 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 4gz6_D | 0.000000000000003 | 161 | 308 | 12 | 152 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 4gz6_D | 0.000000001 | 106 | 233 | 32 | 152 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
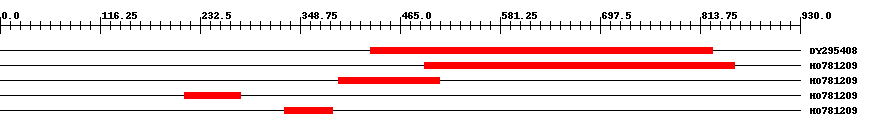 | ||||
| Hit | Length | Start | End | EValue |
| DY295408 | 401 | 431 | 828 | 0 |
| HO781209 | 361 | 494 | 854 | 0 |
| HO781209 | 118 | 394 | 511 | 0 |
| HO781209 | 67 | 215 | 279 | 0.0003 |
| HO781209 | 58 | 331 | 386 | 3.1 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |