

| Basic Information | |
|---|---|
| Species | Gossypium raimondii |
| Cazyme ID | Gorai.002G238200.1 |
| Family | GT41 |
| Protein Properties | Length: 928 Molecular Weight: 103024 Isoelectric Point: 5.7095 |
| Chromosome | Chromosome/Scaffold: 02 Start: 60152274 End: 60159929 |
| Description | Tetratricopeptide repeat (TPR)-like superfamily protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT41 | 362 | 852 | 0 |
| IKPNFSQSLNNLGVVYTVQGKMDAAASMIEKAIIANPTYAEAYNNLGVLYRDAGNITMAVTAYEQCLKIDPDSRNAGQNRLLAMNYINEGDDDKLFEAHR DWGRRFMRLYPQYDSWDNPKDPERPLVIGYISPDYFTHSVSYFIEAPLIYHDYGKYQVVVYSAVVKADAKTNRFRERVVKKGGLWRDIYGIDEKKVASMI RDDKIDILVELTGHTANNKLGTMACRPAPVQVTWIGYPNTTGLPTIDYRITDSLADPPGTKQKHVEELVRLPECFLCYTPSSEAGLVSPTPALSNGFITF GSFNNLAKITPKVLQVWARILCAVPNSRLVVKCKPFCCDSVRQKFLTTLEQLGLESLRVDLLPLILLNHDHMQAYSLMDISLDTFPYAGTTTTCESLYMG VPCVTMAGSVHAHNVGVSLLSKVGLGHLIAKNEDEYVQLALQLASDVTALQNLRASLRDLMSKSPVCDGQNFISGLEATYRGMWRRYCKGD | |||
| Full Sequence |
|---|
| Protein Sequence Length: 928 Download |
| MAWTEKDVNG REKDLIAENG FLKDRQSSPG PSTSTVDVIP PPKAFEGKDA LSYANILRSR 60 NKFVDALAIY NSVLEKDSGC VEAYIGKGIC LQMQNMGRPA FESFAEAIKL DPQNACALTH 120 CGILYKDEGR LVDAAESYQK ALKADASYKP AAECLAIVLT DLGTSLKLAG NTQEGIQKYY 180 EALKIDPHYA PAYYNLGVVY SEMMQYDTAL SCYEKAALER PMYAEAYCNM GVIYKNRGDL 240 ESAIACYERC LAVSPNFEIA KNNMAIALTD LGTKVKLEGD INQGVAYYKK ALYYNWHYAD 300 AMYNLGVAYG EMLKFDMAVV FYELAFHFNP HCAEACNNLG VIYKDRDNLD KAVECYQLAL 360 SIKPNFSQSL NNLGVVYTVQ GKMDAAASMI EKAIIANPTY AEAYNNLGVL YRDAGNITMA 420 VTAYEQCLKI DPDSRNAGQN RLLAMNYINE GDDDKLFEAH RDWGRRFMRL YPQYDSWDNP 480 KDPERPLVIG YISPDYFTHS VSYFIEAPLI YHDYGKYQVV VYSAVVKADA KTNRFRERVV 540 KKGGLWRDIY GIDEKKVASM IRDDKIDILV ELTGHTANNK LGTMACRPAP VQVTWIGYPN 600 TTGLPTIDYR ITDSLADPPG TKQKHVEELV RLPECFLCYT PSSEAGLVSP TPALSNGFIT 660 FGSFNNLAKI TPKVLQVWAR ILCAVPNSRL VVKCKPFCCD SVRQKFLTTL EQLGLESLRV 720 DLLPLILLNH DHMQAYSLMD ISLDTFPYAG TTTTCESLYM GVPCVTMAGS VHAHNVGVSL 780 LSKVGLGHLI AKNEDEYVQL ALQLASDVTA LQNLRASLRD LMSKSPVCDG QNFISGLEAT 840 YRGMWRRYCK GDVPSSRYME MLKKEGVPEG VTNETSKPER VTMSKDTSSV SVESNGFNQA 900 PLSTPNLTTS EDNENQSSQT TNSGKLS* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
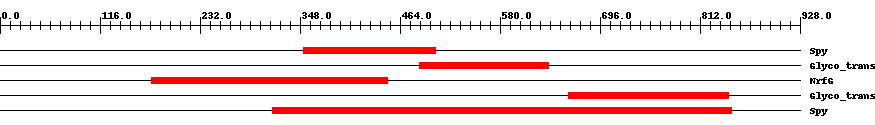 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| COG3914 | Spy | 0.005 | 352 | 505 | 172 | + Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones] | ||
| pfam13844 | Glyco_transf_41 | 3.0e-20 | 487 | 636 | 153 | + Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT, also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. | ||
| COG0457 | NrfG | 5.0e-22 | 176 | 450 | 283 | + FOG: TPR repeat [General function prediction only] | ||
| pfam13844 | Glyco_transf_41 | 1.0e-24 | 659 | 845 | 187 | + Glycosyl transferase family 41. This family of glycosyltransferases includes O-linked beta-N-acetylglucosamine (O-GlcNAc) transferase, an enzyme which catalyzes the addition of O-GlcNAc to serine and threonine residues. In addition to its function as an O-GlcNAc transferase, human OGT, also appears to proteolytically cleave the epigenetic cell-cycle regulator HCF-1. | ||
| COG3914 | Spy | 2.0e-95 | 316 | 848 | 562 | + Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005515 | protein binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| Swiss-Prot | O82039 | 0 | 1 | 927 | 1 | 932 | SPY_PETHY RecName: Full=Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY; AltName: Full=PhSPY |
| RefSeq | XP_002281883.1 | 0 | 1 | 913 | 1 | 911 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002308458.1 | 0 | 1 | 927 | 1 | 934 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002323412.1 | 0 | 1 | 902 | 1 | 907 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002530535.1 | 0 | 1 | 927 | 1 | 930 | o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1w3b_B | 1e-35 | 89 | 440 | 42 | 378 | A Chain A, The Superhelical Tpr Domain Of O-linked Glcnac Transferase Reveals Structural Similarities To Importin Alpha. |
| PDB | 1w3b_B | 6e-27 | 55 | 374 | 75 | 380 | A Chain A, The Superhelical Tpr Domain Of O-linked Glcnac Transferase Reveals Structural Similarities To Importin Alpha. |
| PDB | 1w3b_B | 8e-24 | 219 | 474 | 62 | 338 | A Chain A, The Superhelical Tpr Domain Of O-linked Glcnac Transferase Reveals Structural Similarities To Importin Alpha. |
| PDB | 1w3b_B | 0.0000000000006 | 50 | 271 | 169 | 386 | A Chain A, The Superhelical Tpr Domain Of O-linked Glcnac Transferase Reveals Structural Similarities To Importin Alpha. |
| PDB | 1w3b_A | 1e-35 | 89 | 440 | 42 | 378 | A Chain A, The Superhelical Tpr Domain Of O-linked Glcnac Transferase Reveals Structural Similarities To Importin Alpha. |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| DY295408 | 401 | 429 | 826 | 0 |
| ES841974 | 308 | 616 | 923 | 0 |
| HO781209 | 357 | 496 | 852 | 0 |
| HO781209 | 118 | 392 | 509 | 0 |
| HO781209 | 69 | 211 | 277 | 0.0001 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |