

| Basic Information | |
|---|---|
| Species | Brassica rapa |
| Cazyme ID | Bra009511 |
| Family | GT30 |
| Protein Properties | Length: 454 Molecular Weight: 50674.9 Isoelectric Point: 9.6785 |
| Chromosome | Chromosome/Scaffold: 10 Start: 16995113 End: 16997575 |
| Description | KDO transferase A |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT30 | 56 | 235 | 0 |
| RWPERFGHPSAVRPPGYLVWFHAVSLGEGMAAIPVIRRCNEMKPEMTILMTTTTVSAFEVIKKQLPVGVLHQFAPLDTPVAIDRFLGYWKPNAIVIMENE LWPNLIMSASQLRIPLALLNARMSTKSFKRWSSPLLLPLASLLLSKFSLIAPLSTLQGIHYQLLQAPPFVINFSGDLKYV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 454 Download |
| MNRAEQTSAP SLSGGCAMEL GVLVYRLYRA LTYSASPFLH LHMRWRRLRG LEHSRRWPER 60 FGHPSAVRPP GYLVWFHAVS LGEGMAAIPV IRRCNEMKPE MTILMTTTTV SAFEVIKKQL 120 PVGVLHQFAP LDTPVAIDRF LGYWKPNAIV IMENELWPNL IMSASQLRIP LALLNARMST 180 KSFKRWSSPL LLPLASLLLS KFSLIAPLST LQGIHYQLLQ APPFVINFSG DLKYVVNKFY 240 VSSGTSESIR DLKAELSDMK VWIASSLHRG EEEGVHNLLL QSHPDSVVII VPRHPHHGHQ 300 IAQKLRNDGQ SVALRSRNEK LTSKKTNIYV VDTLGELREF YGVAPIAVIG GSFFPELTGH 360 NVSEAAAAGC AVITGCHVGH FSHMVKAMQQ ENPLSITQVS SELELKEAVE LLMSNPEILE 420 ARRRASKEVY ESLSSCIISN VWNLLNLHIF RGK* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam04413 | Glycos_transf_N | 2.0e-54 | 55 | 234 | 182 | + 3-Deoxy-D-manno-octulosonic-acid transferase (kdotransferase). Members of this family transfer activated sugars to a variety of substrates, including glycogen, fructose-6-phosphate and lipopolysaccharides. Members of the family transfer UDP, ADP, GDP or CMP linked sugars. The Glycos_transf_N region is flanked at the N-terminus by a signal peptide and at the C-terminus by Glycos_transf_1 (pfam00534). The eukaryotic glycogen synthases may be distant members of this bacterial family. | ||
| COG1519 | KdtA | 1.0e-103 | 26 | 430 | 410 | + 3-deoxy-D-manno-octulosonic-acid transferase [Cell envelope biogenesis, outer membrane] | ||
| PRK05749 | PRK05749 | 8.0e-110 | 23 | 429 | 412 | + 3-deoxy-D-manno-octulosonic-acid transferase; Reviewed | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005529 | Interacting selectively and non-covalently with any carbohydrate, which includes monosaccharides, oligosaccharides and polysaccharides as well as substances derived from monosaccharides by reduction of the carbonyl group (alditols), by oxidation of one or more hydroxy groups to afford the corresponding aldehydes, ketones, or carboxylic acids, or by replacement of one or more hydroxy group(s) by a hydrogen atom. Cyclitols are generally not regarded as carbohydrates." [CHEBI:16646, GOC:mah] |
| GO:0005975 | carbohydrate metabolic process |
| GO:0009058 | biosynthetic process |
| GO:0016740 | transferase activity |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| DDBJ | BAB08602.1 | 0 | 18 | 453 | 1 | 432 | 3-deoxy-D-manno-octulosonic acid transferase-like protein [Arabidopsis thaliana] |
| EMBL | CAB82942.1 | 0 | 7 | 453 | 19 | 473 | 3-deoxy-D-manno-octulosonic acid transferase-like protein [Arabidopsis thaliana] |
| RefSeq | NP_195997.2 | 0 | 18 | 453 | 1 | 439 | 3-deoxy-D-manno-octulosonic acid transferase-related [Arabidopsis thaliana] |
| RefSeq | XP_002322900.1 | 0 | 15 | 451 | 2 | 439 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002528642.1 | 0 | 12 | 450 | 1 | 440 | 3-deoxy-d-manno-octulosonic-acid transferase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2xcu_D | 4e-24 | 59 | 430 | 28 | 363 | A Chain A, Crystal Structure Of Escherichia Coli Ybhc |
| PDB | 2xcu_C | 4e-24 | 59 | 430 | 28 | 363 | A Chain A, Crystal Structure Of Escherichia Coli Ybhc |
| PDB | 2xcu_B | 4e-24 | 59 | 430 | 28 | 363 | A Chain A, Crystal Structure Of Escherichia Coli Ybhc |
| PDB | 2xcu_A | 4e-24 | 59 | 430 | 28 | 363 | A Chain A, Crystal Structure Of Escherichia Coli Ybhc |
| PDB | 2xci_D | 4e-24 | 59 | 430 | 28 | 363 | A Chain A, Membrane-Embedded Monofunctional Glycosyltransferase Waaa Of Aquifex Aeolicus, Substrate-Free Form |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| KDO transfer to lipid IVA I | KDOTRANS-RXN | EC-2.4.99.12 | lipid IVA 3-deoxy-D-manno-octulosonic acid transferase |
| KDO transfer to lipid IVA I | KDOTRANS2-RXN | EC-2.4.99.13 | (KDO)-lipid IVA 3-deoxy-D-manno-octulosonic acid transferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
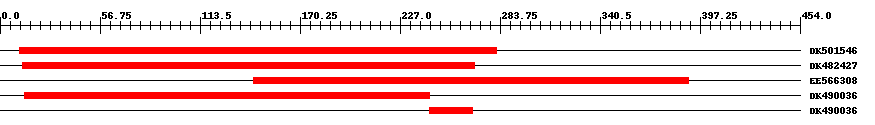 | ||||
| Hit | Length | Start | End | EValue |
| DK501546 | 275 | 11 | 282 | 0 |
| DK482427 | 257 | 13 | 269 | 0 |
| EE566308 | 251 | 144 | 391 | 0 |
| DK490036 | 231 | 14 | 244 | 0 |
| DK490036 | 25 | 244 | 268 | 0.00005 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |