You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002492_00242
You are here: Home > Sequence: MGYG000002492_00242
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
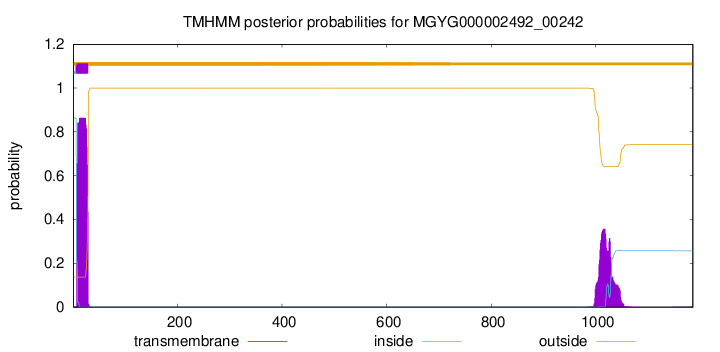
TMHMM annotations
Basic Information help
| Species | Agathobacter rectalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter rectalis | |||||||||||
| CAZyme ID | MGYG000002492_00242 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 256001; End: 259561 Strand: + | |||||||||||
Full Sequence Download help
| MKRLKQRILA GVLAFVMCIS TVNVTAFAQE NETTSEIQEI IAISEAGSEN EVSTENIEIT | 60 |
| DTSVSGNNVE PVEDKPRNKK HKKVANEKVY EGENYNVTFS VTSSWESGYN ANVKVENTGN | 120 |
| DTIQNWYLSF EYDDQITNIW NAEISTHEES QYIIKNADWN QDIVVGGSIE FGISGSTAFN | 180 |
| EFPENFNVVG ENKEVQEEDY SVQYQVDSDW GSGFTGSIQI TNNTDKTLED WVLEFDFERE | 240 |
| ITNIWNAVIE SHEGNHYVIR NAGYNANITA GQSVLFGFNG HGGSANNVAE NCVLYSWEID | 300 |
| TTEYVELSDG KIEKNYLERA IYTNLLLQGL SIDNIRLADD YDEDGLTLSQ EYEYDTNPFS | 360 |
| KDTDEDGLND YEEINLHKTN PIKYDSDEDG MSDGTEIACG LNPLSSDTDG NGVIDSQEIV | 420 |
| TQAVRIDTVE QYQLQEVGTL PNIQITGKGD YSQKIYATAL ENDATIMDID CLVGTAFDFI | 480 |
| HDEDLSFENS QLTFTISNEI LKKNKLTDLA IAWYNEEENA LELLDTTHNM DNCTISAEVS | 540 |
| HYSTYMVVSV PDYFFNIDWE NEDSIIEAGK ADVVFVIDTT GSMGNEIQNV KNNIETVVSS | 600 |
| LEENKVDIRL GLVEYRDIYA DGIGSTKSYD WYTSVSSFKS ELATLGVSGG GDTPESVVDA | 660 |
| LYCARNMEYR TGVKKYVILL TDANYKNGTS VDSGATLTDE IRRLVEEELV VSVVTTPSYY | 720 |
| STYNSLVSQT DGVTANINQN FASALAPLIT KMGEQVNQGC WIRLSNGSVV SLDKDPTLGD | 780 |
| ETIDTDDDGI PDIIELKSSY KVHAYNPYTK KMQEIDTWSF YSNPSKRDTD GDTLSDIDDL | 840 |
| QPTKYDTVVI KENDSSIAFN TGRTWYNISC TSFDYLDNLM QMVDGKVDNP IPIEQFRQII | 900 |
| QNVANNEKQA FTIEELTYIA IMNNEGSKLY MHNLSSVTRE NVFQKIAGRE SRYYKHSGIW | 960 |
| WNENWSEVPK GTESGFFKGT VLSEADINLS WEIYCVCDVY TVLTTVAQVG ALVIAIVVVA | 1020 |
| EVTPVVLANI QGLAYYVKTF GIVQGIQMYR YLGIQNLPNG VISWLQMDMA DGDSSVDDVA | 1080 |
| LAIQKESVEK KLDTYLLNKD HPVGGSKANW FEKALGFTKE NAGQLAKQIV FDENVAVQTA | 1140 |
| VTEHGIKYNQ LISIVGANGK KIDVVFVWIK NNDGFVRLVT AIPTKK | 1186 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM2 | 201 | 293 | 1.9e-26 | 0.9207920792079208 |
| CBM2 | 97 | 190 | 2.3e-23 | 0.9405940594059405 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00553 | CBM_2 | 1.96e-24 | 202 | 294 | 3 | 96 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 7.67e-21 | 207 | 294 | 1 | 88 | CBD_II domain. |
| pfam00553 | CBM_2 | 6.01e-20 | 96 | 191 | 2 | 99 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 1.43e-19 | 102 | 190 | 1 | 90 | CBD_II domain. |
| smart00327 | VWA | 9.57e-13 | 572 | 703 | 1 | 128 | von Willebrand factor (vWF) type A domain. VWA domains in extracellular eukaryotic proteins mediate adhesion via metal ion-dependent adhesion sites (MIDAS). Intracellular VWA domains and homologues in prokaryotes have recently been identified. The proposed VWA domains in integrin beta subunits have recently been substantiated using sequence-based methods. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACR74072.1 | 0.0 | 1 | 1186 | 1 | 1186 |
| CBK93270.1 | 8.41e-106 | 1 | 840 | 32 | 872 |
| ADL33479.1 | 2.38e-58 | 95 | 279 | 68 | 253 |
| CDZ23468.1 | 3.76e-58 | 90 | 687 | 35 | 730 |
| QEH69368.1 | 6.36e-55 | 90 | 894 | 104 | 1080 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT9_A | 2.57e-16 | 94 | 176 | 576 | 658 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
| 2RTT_A | 3.26e-13 | 208 | 291 | 12 | 97 | Solutionstructure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor [Streptomyces coelicolor] |
| 6QFS_A | 2.17e-12 | 103 | 190 | 11 | 100 | ChainA, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_B Chain B, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_C Chain C, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_D Chain D, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_E Chain E, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_F Chain F, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_G Chain G, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_H Chain H, Exoglucanase/xylanase [Cellulomonas fimi] |
| 1EXG_A | 1.15e-11 | 103 | 190 | 15 | 104 | ChainA, EXO-1,4-BETA-D-GLYCANASE [Cellulomonas fimi],1EXH_A Chain A, EXO-1,4-BETA-D-GLYCANASE [Cellulomonas fimi] |
| 3NDY_E | 3.68e-06 | 202 | 279 | 5 | 84 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_E The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36909 | 1.02e-10 | 208 | 354 | 42 | 204 | Chitinase C OS=Streptomyces lividans OX=1916 GN=chiC PE=2 SV=1 |
| P11220 | 1.74e-10 | 204 | 354 | 38 | 204 | Chitinase 63 OS=Streptomyces plicatus OX=1922 GN=chtA PE=1 SV=2 |
| P18126 | 2.30e-09 | 99 | 176 | 34 | 111 | Endoglucanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celB PE=1 SV=1 |
| P07986 | 4.98e-09 | 103 | 190 | 389 | 478 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| P27033 | 9.20e-09 | 99 | 177 | 41 | 118 | Endoglucanase C OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celC PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000331 | 0.998896 | 0.000247 | 0.000178 | 0.000168 | 0.000152 |