You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000932_00119
You are here: Home > Sequence: MGYG000000932_00119
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
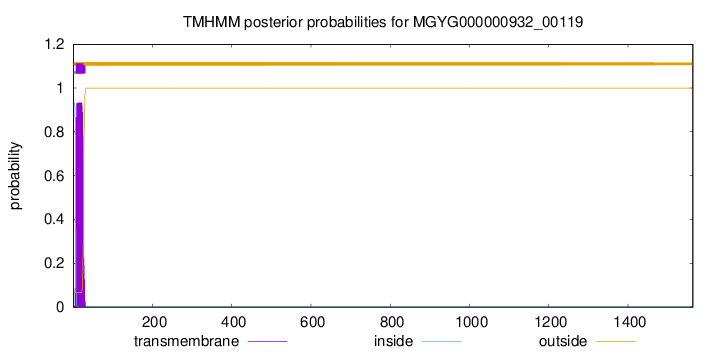
TMHMM annotations
Basic Information help
| Species | UMGS577 sp900543525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; UMGS577; UMGS577 sp900543525 | |||||||||||
| CAZyme ID | MGYG000000932_00119 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20574; End: 25268 Strand: + | |||||||||||
Full Sequence Download help
| MKFKRICAAV LALLILSTTF PAVQAARLPA LPSGPYTLAP ESPAIIGLKC LDAQNADKWS | 60 |
| VESGLAAGAA VYGDADIHFT DVDRELEGCT WIRTADASAQ YKGEGALAAF TAGKSGRLYI | 120 |
| GLPAGVEKPG WMEAYTASPL TLRTASGSFS LYQKACETGD RVELFSINEA HNYTLLYKTD | 180 |
| VIDEAPPVPN VTKFWTVEWD AGWYRMETDL QIGRQIYSFD YDSDGSEVKT QNPDHFILTA | 240 |
| LPEKYIGCDY IVTRANRARD LYFFAEQNID VYAAYDSRQS KPAQLSDWTA TGDSMKIQDE | 300 |
| TSYNIVQKSF PMGETVLLKA TGGVGNSVRN TFYMILLTSG ETVSKALTSN PVVAQGEKPE | 360 |
| DSAADQNYRY YLNEVFNLYE GSGLPNGYTG SGAAVADANE LPDADAVTEK SFAAGENMAL | 420 |
| NVPYSYNTIS YSGSPVDGSI DTYWQNAKDD FPGILTIDLQ GQCRIDKITL KLRDYWADRT | 480 |
| QNLELQTSTD GKSFTTLIAA TDYLFEKSKQ NVLDLSFPET TARYVRMIGN SNTGWNTVQL | 540 |
| SEIEVYGPAQ TIEIEPEAPT ESRFTDRCAV IEKTGKEDAV LAKAFDEAVS GKLVFEAKVR | 600 |
| SSADNQTMAI PVMEDSSKNP ALALYFGEDG YIKSVNSTEE ISVAPYTAKT WYTIKLVIDT | 660 |
| RSGKYDLWID HLLKGRDLSL YRGAKDIQTI RFGVINPSCG SLSVDNLRLY DNPEVYTVED | 720 |
| RFNHLETGSL PQDGWSCSEN SSAAVAEVPF PSDKSLLIQN TGDAAKAVRS FTPITGDVTV | 780 |
| EAKIKASGSG WVSVPIITDK AGRVAVKVAA YRNSFYICSG NNWVYICSQE VPNNYYPADN | 840 |
| WYFIKVVLNT YTNRYDFYID GAKRYSGASF AADVDEVSRL VFAAEQENTL YIDNVKVYDS | 900 |
| ASLARGLMPK ENLFNVKAFG AKGDGVTDDT EAIAKAIDAA AGTDGTVLLE NGVFYTGQIT | 960 |
| LESDMTLFID ASATILANPD RNVYQKVIPS RGYNGNRQLG RGIIYGESLS NTRITGGGTI | 1020 |
| QGNGFYAFNE NDPSNQRPCT IYITLSNDIS VENINMVQSP FWTLVPYESS GFTVRNVSIT | 1080 |
| NHTAPNRDGI DPVNTSNMTV ENCYIIAGDD AFCPKSGNDI PSCNIDVREV YMQSYCNGIK | 1140 |
| FGTDSYDAFK NYTIEDITMK CVGLSGITLQ SSDGAEIENI TFRRIDMNDV DNVLSFMVGN | 1200 |
| RCRTPIDATA PRRLGYIRNI VVEDLNYTNP MQVPYSHKDE DVHEAMIIGL DPAKNTLNDG | 1260 |
| EAHRISNILF KNAKLEMPGG AASVPAFSGG IGGGYPEHTG LGSSTGWAYT IRWADNVRFE | 1320 |
| NCKNTLQNPD VRAEIATQDY TDTPVTAPLR AAYVMAVPEI HLPTGTAASG LKLPKAVSVL | 1380 |
| LKDGSVAKAA AAEWKSDPGY DSDTPGVYTF EAVIAANDQI LDADKLTAKA TVRVLEQTAY | 1440 |
| DALKAAEVDM QIDRLGAVVS PEQEARIQSV RAEFKKLSDA QKSMVKQLPV LEAAEKAIAL | 1500 |
| LKNPYQKGDL DRDRRVTVSD VVALRQMIVT GSATDEDLRL GDMDEDDRLT VSDVVLLRQR | 1560 |
| IVKG | 1564 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 941 | 1246 | 6.5e-57 | 0.84 |
| CBM32 | 436 | 543 | 2.1e-18 | 0.7983870967741935 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.05e-38 | 914 | 1203 | 83 | 411 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02188 | PLN02188 | 7.01e-17 | 913 | 1192 | 36 | 312 | polygalacturonase/glycoside hydrolase family protein |
| pfam00295 | Glyco_hydro_28 | 1.86e-13 | 1004 | 1190 | 48 | 239 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN02218 | PLN02218 | 5.53e-13 | 902 | 1192 | 56 | 347 | polygalacturonase ADPG |
| pfam00754 | F5_F8_type_C | 5.96e-13 | 436 | 542 | 18 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOZ98256.1 | 2.15e-61 | 910 | 1339 | 22 | 444 |
| QES88576.1 | 2.44e-57 | 892 | 1335 | 150 | 591 |
| SOE23521.1 | 1.68e-56 | 913 | 1332 | 25 | 437 |
| QUI22115.1 | 1.03e-48 | 913 | 1339 | 2 | 421 |
| AWG23677.1 | 2.07e-43 | 914 | 1339 | 28 | 471 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 3.05e-21 | 915 | 1228 | 29 | 384 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 1BHE_A | 2.14e-11 | 935 | 1190 | 40 | 304 | ChainA, POLYGALACTURONASE [Pectobacterium carotovorum] |
| 2UVE_A | 1.65e-07 | 891 | 1190 | 134 | 490 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 1.05e-22 | 919 | 1225 | 68 | 395 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| Q8RY29 | 5.85e-14 | 902 | 1229 | 56 | 382 | Polygalacturonase ADPG2 OS=Arabidopsis thaliana OX=3702 GN=ADPG2 PE=2 SV=2 |
| P35336 | 2.18e-13 | 915 | 1283 | 91 | 436 | Polygalacturonase OS=Actinidia deliciosa OX=3627 PE=2 SV=1 |
| Q94AJ5 | 3.25e-12 | 904 | 1192 | 42 | 332 | Probable polygalacturonase At1g80170 OS=Arabidopsis thaliana OX=3702 GN=At1g80170 PE=1 SV=1 |
| P48978 | 8.11e-12 | 848 | 1229 | 35 | 412 | Polygalacturonase OS=Malus domestica OX=3750 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
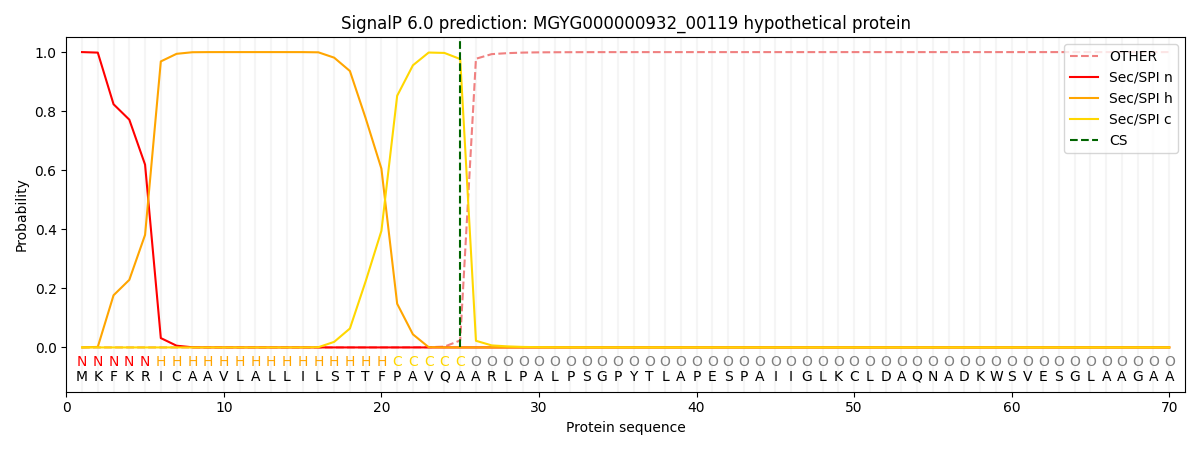
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000233 | 0.999067 | 0.000191 | 0.000183 | 0.000169 | 0.000147 |