You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001183_00465
You are here: Home > Sequence: MGYG000001183_00465
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900551755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900551755 | |||||||||||
| CAZyme ID | MGYG000001183_00465 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 489530; End: 493771 Strand: - | |||||||||||
Full Sequence Download help
| MNIFKQIIAI GVSATFTLQA MPLTAYGEDE TTNNQLTIAE ISSTNTLMSS EYWANHRIST | 60 |
| NTSDDTYIAS YTNNAGQYYS SSLDKAFDGN WSTHWETGNG NTHNYVDITF NKAVTIDRML | 120 |
| YATRQDGAKP KGYPENLTIY SYNSSTEQWD EVANGTSTKT GGYVLITLPQ ATTFEKLRFD | 180 |
| FTQAYNGWAS ASEFVFLRPD NTVIDGSINI NGTPLVGETL TAIPNLTVGD NNNINYQWQY | 240 |
| SDNGVDYYNI ENATNSTYKI TEKKNYFRVN VTDSNKNYYG TLSSNSLDYS KAITTGSPQA | 300 |
| NATLTASIYN VDKKFTCKWQ ISDSQDGTYT DIEKATAISY TLTESDVNKY IKAILTLTET | 360 |
| GETLETEPKL ISADGTYNVY TSDYVNLSEL PSSALIQSSV GYDKLRYNEN IDGNTISLLV | 420 |
| NNTVKKFSKG LSAHAAATLV YDVSFFTENY QYNKLTAWLG VDKSTQLVGD GVKFYVYTSN | 480 |
| DNSNWTLVES TDALMSNSEA VYIDINIKNV KYVKFYISSI SGNSNDHSVI AGAKLSKSDY | 540 |
| TEGSDNYDFI KTISQYDKEL RTYENSNSYE SLVNNEDYRK LLYQRTFVNY VDYDVLQSIA | 600 |
| HDSDSNKKAL EWFMNDFEAL DYYINGGPIV KGTSYQRSVN ALINLYTAYS GDMDNSLYKR | 660 |
| LLIALALVNS EPITSWVDSS TVSDPVRRYG IFKKLYNNGY LVNNIFENLS VAEMKWVVNN | 720 |
| RISDEEIEWL NYFLRKDYYK NVEADSDSIY DSIVNNKFVN PYNYITYGNG YNYSLDKYYS | 780 |
| EENKAKWQEK YHLVNTDDTD DDNFDISVTY ESGHPRLWIV FEEGSVCGGI AGTGANFSTV | 840 |
| FGVPSMLVGQ PGHAAYFRYR HDPGDNGQYK LGIWELWNDV YGWSKTWIAG GLINGWGTES | 900 |
| WCSAYQGNYL LLVQAALNDY DNYIKAEDFV KMASTDTTDT DKVIELCEKA LSVQNFNVDA | 960 |
| WELLINAYVS AGKSEDEFLN LAQRVTTDLK YYPHPMRDIV VNLIQPNIST SSLLADCELY | 1020 |
| LNSSLTEATK ATKNDTLQPS YCKDVANYIL GNENHTLASF SFSGDSANKI MLTDTYKDNG | 1080 |
| NELLYSIDGG TTWINAGMVN EYQLSNDEIS KITADNDILV KLQGSSNYYT IDIKTSTAPK | 1140 |
| SLFSNDNENK IFGTTDTMEW STDKITWESF NNTLFTGDKT VYVRTKANGT TLPSDSVLFN | 1200 |
| FTADSSSGER TYITIDRITV DKYSSAHQND KAENTIDGNI NTIWHSEYSN STSTDSERYI | 1260 |
| SYKFNKPIYL SAVDYTPRQS GQNGIFTTCE IYTSLNGTDW KLATTATGLA NNASTKSIDL | 1320 |
| DTPVYANYVK VVGKEAIGNF GSASMIEFFE NTTIGKNSAD LNNDGENNSI DVATLLKYVS | 1380 |
| EMTNVTIDES KTDINGDGKT DIADVIALAK YIE | 1413 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 388 | 535 | 3.8e-20 | 0.9701492537313433 |
| CBM32 | 1224 | 1348 | 3.3e-18 | 0.9032258064516129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.88e-23 | 383 | 536 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam00754 | F5_F8_type_C | 4.87e-15 | 1224 | 1348 | 6 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| smart00776 | NPCBM | 1.29e-10 | 381 | 536 | 1 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| cd14256 | Dockerin_I | 3.10e-08 | 1360 | 1412 | 3 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00754 | F5_F8_type_C | 7.97e-08 | 84 | 194 | 15 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35280.1 | 1.94e-196 | 383 | 1350 | 47 | 987 |
| AQW23377.1 | 3.09e-194 | 383 | 1350 | 47 | 987 |
| ATD49073.1 | 3.57e-194 | 383 | 1350 | 52 | 992 |
| SQG16429.1 | 1.46e-143 | 356 | 1350 | 112 | 1098 |
| VED84782.1 | 1.46e-143 | 356 | 1350 | 112 | 1098 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JND_A | 3.57e-22 | 68 | 231 | 72 | 241 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 6.37e-22 | 68 | 231 | 72 | 241 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JFS_A | 2.15e-21 | 68 | 231 | 54 | 223 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2V72_A | 1.07e-12 | 1224 | 1350 | 21 | 143 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 4LKS_A | 3.43e-08 | 1201 | 1349 | 19 | 165 | Structureof CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LKS_C Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LQR_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124],4P5Y_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 2.79e-07 | 1220 | 1300 | 514 | 594 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P0DTR4 | 1.19e-06 | 1232 | 1349 | 530 | 644 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q0TR53 | 5.05e-06 | 1217 | 1331 | 630 | 741 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 5.05e-06 | 1217 | 1331 | 630 | 741 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
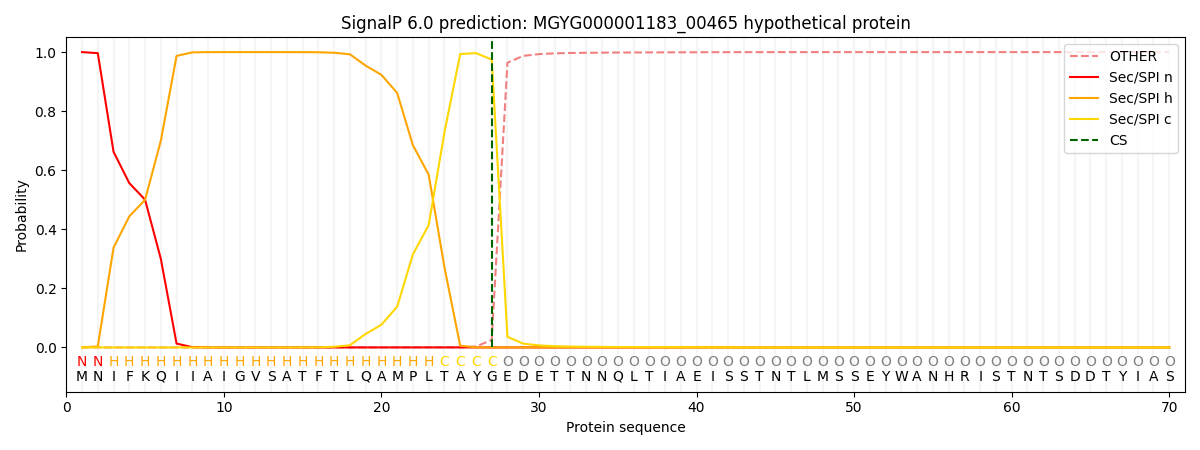
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998961 | 0.000223 | 0.000180 | 0.000169 | 0.000145 |
