You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004288_00531
You are here: Home > Sequence: MGYG000004288_00531
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
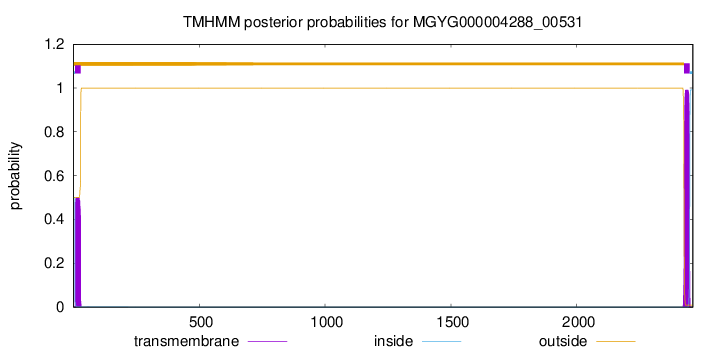
TMHMM annotations
Basic Information help
| Species | Faecalicoccus pleomorphus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Faecalicoccus; Faecalicoccus pleomorphus | |||||||||||
| CAZyme ID | MGYG000004288_00531 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 109041; End: 116426 Strand: - | |||||||||||
Full Sequence Download help
| MKRHQKAILA SVVTASQAAT LLNIPVTVMA QEPDVSRTDR VAKNEALQPK SGYTFIGAPV | 60 |
| AMPEVQNNIV TVRYANGEKA KITFLENNLF RFDMEVDGQD GSFKDYADPN DVSHTGRIVQ | 120 |
| QPDSSDEYTK PTPTVGETQD AYSISNDAII LEVSKTDARM TLKRTDGTVV WQEAAPIQYK | 180 |
| TGSTIQSFVK NEGENFYGGG TQNGRFVHTG ESIKIENLNN WVSGGVASPN PFYWSTNGYG | 240 |
| VLRNTFKKGE YDFGKTQADV VSTSHTEKRL DAYYFVGDTP ISLLQSYFKV TGNPALFPEE | 300 |
| AFYLGHLNCY NRDEWTEGGG TPLETINDSS KYQETNNGGK IKRGGVLETL NGTNETDYKF | 360 |
| SARGVIDQYD KYDMPLGWFL PNDGYGCGYG QDDTNLDNNI ANLKSFTDYA NSKGVGTGLW | 420 |
| TQSDLTPNPN QPIHLQRDFE KEVYQGGIRT LKTDVAWVGA GYSFGLNGIS KAYDIITKAD | 480 |
| TRNRPTIVTL DGWAGTQRYG GIWTGDQTGG NWEYIRFHIP TYIGQSLSGN PNVSSDVDGI | 540 |
| FGGSSLIQTR DIQWKSFTTM MLDMDGWGSY PKKPYVFGED TTSINRMYLK LRSQLMPYIY | 600 |
| STAYSSANLG EDGEKGKPQV RAMFLEFPED ANAYGKNVQY QFMLGKNLLV APVYQNTAAD | 660 |
| GQGNDIRNGI YLPDSNQVWI DYFTGKQYQG GSTLNNFDAP IWKLPLFVKN GAILPMTEAH | 720 |
| NNGFAKSETN TKGVDKSKRV VEFYPAGETD YTLYEDAGNT VDNTNTSEVN YGSAVTTHFT | 780 |
| SKVTGEKAVL KAEASQGSYV GYDANKETQF VVNVSEKPSS IQATVGANDA NLTEVHSQKE | 840 |
| FDEATGNVYY YNESPDLNRF ATPESDFASV KITTTPKLYV KFAKTDVSAN AIELSLEGFK | 900 |
| NDGDLDKNVL NKDLKAPANV RAPEESITPD TIHLQWDAVE NATSYEVEAD GVIQTGFTEP | 960 |
| EFDHTDLAYH SKHTYRVRAR NKDGYSQWSE TLEVQSAQDP YRNVPKDIQV TWEYGDAWGK | 1020 |
| LANILDFDTG TMFHSTKAVQ DGQAMIYDLK KIYDLDRIEY TPRLDNKGNG TVQKMDVYTS | 1080 |
| LDGTNWTLAY DSSKNPEWTY SKDMSDPDTK TMDMTGQKAR YIKYVVKKSV GGFFSAAELQ | 1140 |
| PYKVDGTEGY IVGDTNNDGT VDDNDVVQID NYVGLEEGDP TWDQVKRSDW NDNKYFDAAD | 1200 |
| IAQTTSRLGK GIAKTGGDPK GSLVIEPDKA HYSAGEKVTL SIQGISMENV YALGGKIPYN | 1260 |
| SEDLQFVSVS SDIATGDQRE FVFDRIAYQN QEHPDKNRNV NFAYSNLGDQ PMLSGTNRLA | 1320 |
| TVTFTAKKDL DIKASDFNLS DFMFVSNNMN YIDPLKPSED LPEPETESKI QVLSVEGQDE | 1380 |
| SVLQPGMGVD KLIDGAWGSD ANRFEFKWGN DEASVPARLP YWIRFNFGEI KKVSDVIIHV | 1440 |
| RVDGNRINGG ALKDFDLYRI KNGVEQKIGS YVIDKTEDEN AYVIHFADPI QAEGFKLNAK | 1500 |
| TSQAGQIFKL NIDEVEFRQN AQTPIEDITL DEANPTTLNV GDLKGFKASV APDTATNKLI | 1560 |
| KIESSDPEVL EVIRTTRDDH YEYTLHALKP GTVTLTITSN GQTVQGEAIT RQVEITVNEA | 1620 |
| VTSKEELESK IEEANAILAN ENLYTPSSLD VLKNALKLAQ NVEDKKDATQ TEVNAQVVQL | 1680 |
| YKAIQQLEYR GSNTAQPDSE NPIVVDPANV TATTSASEGP VENAFDGDQA TYWHSGWEQG | 1740 |
| TEKLPQSVTV DLADEYDVEQ VNYLPRQGSR NGDIIKYQIE TSEDGETFKP VVVGTIENDG | 1800 |
| SSIVERTQPH KIKFDKTKAR YVRITALESL GNQNNLYASA AEFSIFGALH SDAIPATSIE | 1860 |
| LDKTELKLDA GNTETVTATI LPEDTTDTIT WESSDPSIAS VEALGRRRVR RDANASSNTS | 1920 |
| TVTVTALKAG TVTITAKAND TVSKTLTVTV ENPKQEQLAE LIEQAKGVKY ENAALQDYLT | 1980 |
| SEIEKATQAL DQDQDTLQNA YYALAGALSE IEEIQKDIDQ LNSFSDIDDS RYVPNEQYQM | 2040 |
| FKEQLQLAQN LLSDPVQNKE LIKTHVDALK KAFAQLEELD REKLSNAIQE AEKVNLQDCV | 2100 |
| EDDALSAFKT ALENAKAANP QSNEEIDTLV NTLLEAQAGL HFKDENLATD DQKESIQHAL | 2160 |
| DVLKNLDLEL YSEQDGKTIK AAISDGEDAL ANKDLTKEEA SKVIDDLAKA LSLEPKEVTP | 2220 |
| EEEPASSSQK EVIAKALDIL KGLDLNLYDT KAQDVIQETI EEAQKAADND QLTKEEAANV | 2280 |
| LEHLANALSI QPKEDTPKDE KATSTQKEVI EKALDILKDL DLNKYEAKDA DLIKEAIEAT | 2340 |
| QKALENKELT YAEATDTIEK IAQALLVTPK ETEQPDPSDN ENPNTNTPDH GQNNSTDTGS | 2400 |
| NGSNTVIVNP TDSSKSKGNT KTGVHSFVGL FAALATSAAA TAGTVTLLKN KKIRIRRKGK | 2460 |
| K | 2461 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH31 | 467 | 714 | 2.6e-58 | 0.5339578454332553 |
| CBM32 | 1712 | 1844 | 1.7e-26 | 0.9112903225806451 |
| CBM32 | 1016 | 1139 | 1.4e-16 | 0.8709677419354839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06596 | GH31_CPE1046 | 1.85e-171 | 290 | 685 | 1 | 334 | Clostridium CPE1046-like. CPE1046 is an uncharacterized Clostridium perfringens protein with a glycosyl hydrolase family 31 (GH31) domain. The domain architecture of CPE1046 and its orthologs includes a C-terminal fibronectin type 3 (FN3) domain and a coagulation factor 5/8 type C domain in addition to the GH31 domain. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| COG1501 | YicI | 2.17e-68 | 137 | 786 | 99 | 735 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| pfam01055 | Glyco_hydro_31 | 1.04e-60 | 273 | 714 | 1 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| cd06603 | GH31_GANC_GANAB_alpha | 1.66e-39 | 484 | 716 | 213 | 428 | neutral alpha-glucosidase C, neutral alpha-glucosidase AB. This subgroup includes the closely related glycosyl hydrolase family 31 (GH31) isozymes, neutral alpha-glucosidase C (GANC) and the alpha subunit of heterodimeric neutral alpha-glucosidase AB (GANAB). Initially distinguished on the basis of differences in electrophoretic mobility in starch gel, GANC and GANAB have been shown to have other differences, including those of substrate specificity. GANC and GANAB are key enzymes in glycogen metabolism that hydrolyze terminal, non-reducing 1,4-linked alpha-D-glucose residues from glycogen in the endoplasmic reticulum. The GANC/GANAB family includes the alpha-glucosidase II (ModA) from Dictyostelium discoideum as well as the alpha-glucosidase II (GLS2, or ROT2 - Reversal of TOR2 lethality protein 2) from Saccharomyces cerevisiae. |
| cd06589 | GH31 | 9.52e-38 | 351 | 589 | 15 | 265 | glycosyl hydrolase family 31 (GH31). GH31 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. In most cases, the pyranose moiety recognized in subsite -1 of the substrate binding site is an alpha-D-glucose, though some GH31 family members show a preference for alpha-D-xylose. Several GH31 enzymes can accommodate both glucose and xylose and different levels of discrimination between the two have been observed. Most characterized GH31 enzymes are alpha-glucosidases. In mammals, GH31 members with alpha-glucosidase activity are implicated in at least three distinct biological processes. The lysosomal acid alpha-glucosidase (GAA) is essential for glycogen degradation and a deficiency or malfunction of this enzyme causes glycogen storage disease II, also known as Pompe disease. In the endoplasmic reticulum, alpha-glucosidase II catalyzes the second step in the N-linked oligosaccharide processing pathway that constitutes part of the quality control system for glycoprotein folding and maturation. The intestinal enzymes sucrase-isomaltase (SI) and maltase-glucoamylase (MGAM) play key roles in the final stage of carbohydrate digestion, making alpha-glucosidase inhibitors useful in the treatment of type 2 diabetes. GH31 alpha-glycosidases are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBK23937.1 | 0.0 | 2 | 2228 | 4 | 2209 |
| QJA02122.1 | 0.0 | 1 | 2215 | 1 | 2224 |
| BCT46261.1 | 0.0 | 56 | 2076 | 43 | 2115 |
| QIK85989.1 | 0.0 | 50 | 2289 | 41 | 2257 |
| QBJ76643.1 | 0.0 | 51 | 2211 | 52 | 2204 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 6.55e-266 | 54 | 1002 | 35 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 3.48e-265 | 54 | 1002 | 35 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 9.47e-265 | 54 | 1002 | 35 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 4LKS_A | 1.07e-30 | 1712 | 1847 | 31 | 166 | Structureof CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LKS_C Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LQR_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124],4P5Y_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine [Clostridium perfringens ATCC 13124] |
| 6JR8_A | 1.51e-30 | 130 | 865 | 90 | 828 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9F234 | 4.34e-31 | 112 | 760 | 69 | 713 | Alpha-glucosidase 2 OS=Bacillus thermoamyloliquefaciens OX=1425 PE=3 SV=1 |
| Q9P999 | 2.92e-29 | 143 | 762 | 56 | 662 | Alpha-xylosidase OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=xylS PE=1 SV=1 |
| Q9FN05 | 2.40e-27 | 270 | 762 | 332 | 822 | Probable glucan 1,3-alpha-glucosidase OS=Arabidopsis thaliana OX=3702 GN=PSL5 PE=1 SV=1 |
| B9F676 | 9.70e-25 | 270 | 762 | 330 | 820 | Probable glucan 1,3-alpha-glucosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os03g0216600 PE=3 SV=1 |
| Q9BE70 | 1.25e-24 | 469 | 716 | 405 | 636 | Neutral alpha-glucosidase C (Fragment) OS=Macaca fascicularis OX=9541 GN=GANC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
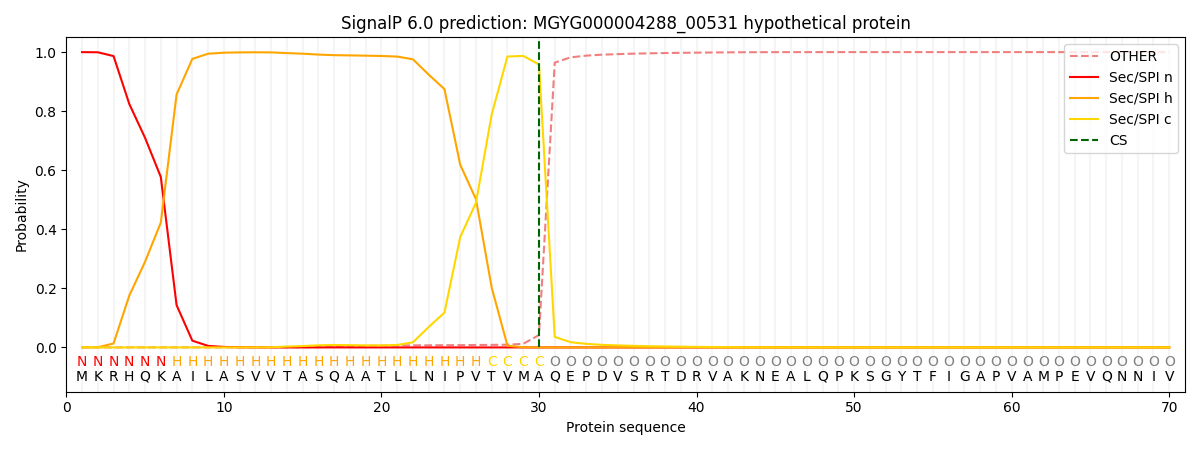
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000670 | 0.998413 | 0.000259 | 0.000283 | 0.000198 | 0.000167 |