You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003370_00337
You are here: Home > Sequence: MGYG000003370_00337
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
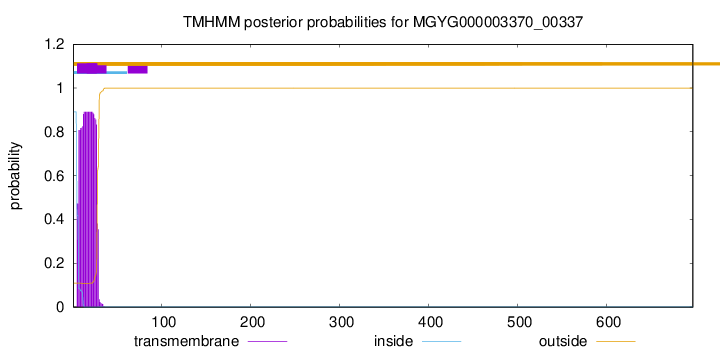
TMHMM annotations
Basic Information help
| Species | Streptococcus mitis_AC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus mitis_AC | |||||||||||
| CAZyme ID | MGYG000003370_00337 | |||||||||||
| CAZy Family | CBM40 | |||||||||||
| CAZyme Description | Sialidase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16261; End: 18354 Strand: - | |||||||||||
Full Sequence Download help
| MNKRGLYSKL GISVVGISLL MGVPTLIHAN ELNYGQLSIS PIFQGGSYQL NNKSIDISPL | 60 |
| LLDKLSGESQ TVVMKFKADK PNSLQALFGL SNSKAGFKNN YFSIFMRDSG EIGVEIRDAQ | 120 |
| KGINYLFSRP ASLWGKHKGQ AVENTLVFVS DSKDKTYTMY VNGIEVFSET VDTFLPISNI | 180 |
| NGIDKATLGA VNREGKDHYL AKGSIDEISL FNKAISDQEV STIPLSNPFQ LIFQSGDSTQ | 240 |
| ANYFRIPTLY TLSSGRVLSS IDARYGGTHD SKSKINIATS YSDDNGKTWS EPTFAMKFND | 300 |
| YEEQLVYWPR DNKLKNSQIS GSASFIDSSI VEDKKSGKTI LLADVMPAGI GNNNANKADS | 360 |
| GFKEINGHYY LKLKKDGDND FRYTVRENGV VYDERTNKPT NYTVNDKYEV LEGGKSLTVE | 420 |
| QYSVDFDSGS LRETHNGKQV PMNVFYKDSL FKVTPTNYIA MTTSQNRGES WEQFKLLPPF | 480 |
| LGEKHNGTYL CPGQGLALKS SNRLIFATYT SGELTYLISD DSGQTWKKSS ASIPFKNATA | 540 |
| EAQMVEMRDG VIRTFFRTTT GKIAYMTSRD SGETWSEVSY IDGIQQTSYG TQVSAIKYSQ | 600 |
| LIDGKEAVIL STPNSRNGRK GGQLVVGLVN KEDDSIDWKY HYDIDLPSYG YAYSAITELP | 660 |
| NHHIGVLFEK YDSWSRNELH LSNVVQYIDL EINDLTK | 697 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 232 | 676 | 4.5e-76 | 0.9415204678362573 |
| CBM40 | 44 | 225 | 3.1e-64 | 0.9720670391061452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4409 | NanH | 0.0 | 1 | 697 | 1 | 728 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam02973 | Sialidase | 1.97e-72 | 42 | 227 | 1 | 188 | Sialidase, N-terminal domain. |
| cd15482 | Sialidase_non-viral | 1.06e-53 | 230 | 684 | 1 | 338 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 2.18e-13 | 459 | 666 | 82 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13385 | Laminin_G_3 | 7.18e-07 | 95 | 221 | 39 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBP09272.1 | 0.0 | 1 | 697 | 1 | 697 |
| ARD35268.1 | 0.0 | 1 | 697 | 1 | 697 |
| AZT87744.1 | 0.0 | 1 | 697 | 1 | 697 |
| AFC95267.1 | 0.0 | 1 | 697 | 1 | 697 |
| ACO22908.1 | 0.0 | 1 | 697 | 1 | 697 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VW0_A | 0.0 | 1 | 697 | 1 | 697 | Crystalstructure of the NanB sialidase from Streptococcus pneumoniae [Streptococcus pneumoniae],2VW1_A Crystal structure of the NanB sialidase from Streptococcus pneumoniae [Streptococcus pneumoniae],2VW2_A Crystal structure of the NanB sialidase from Streptococcus pneumoniae [Streptococcus pneumoniae],4FOQ_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-aminoethanesulfonic acid [Streptococcus pneumoniae],4FOV_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 3-Cyclohexyl-1-propylsulfonic acid [Streptococcus pneumoniae],4FOW_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 3-ammoniopropane-1-sulfonate [Streptococcus pneumoniae],4FOY_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-(Benzylammonio)ethanesulfonate [Streptococcus pneumoniae],4FP2_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2[(Cyclohexylmethyl)ammonio]sulfonate [Streptococcus pneumoniae],4FP3_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(Furan-2-ylmethyl)ammonio]sulfonate [Streptococcus pneumoniae],4FPC_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(4-Chlorobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPE_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(4-Methoxybenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPF_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-Chlorobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPG_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-Hydroxybenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPH_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-Fluorobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPJ_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-Methoxybenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPK_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-methylbenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPL_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3,4-dichlorobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPO_B Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-chloro-4-methoxybenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FPY_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(3-Bromobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae],4FQ4_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 2-[(4-Fluoro-3-methylbenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae] |
| 4XE9_A | 0.0 | 39 | 696 | 1 | 658 | Crystalstructure of the NanB sialidase from Streptococcus pneumoniae in complex with Optactin [Streptococcus pneumoniae TIGR4],4XHB_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with pentanediol and CHES [Streptococcus pneumoniae TIGR4],4XHX_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with Optactin and 2-[(3-Chlorobenzyl)ammonio]ethanesulfonate [Streptococcus pneumoniae TIGR4],4XIK_A Crystal structure of NanB sialidase from streptococcus pneumoniae in complex with DMSO [Streptococcus pneumoniae TIGR4],4XIL_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with Optactin at pH 7.4 in PBS with MPD as the cryoprotectant [Streptococcus pneumoniae TIGR4],4XIO_A Crystal structure of apo NanB sialidase from streptococcus pneumoniae at pH 8.0 with MPD as the cryoprotectant [Streptococcus pneumoniae TIGR4],4XJ8_A Crystal structure of apo NanB sialidase from streptococcus pneumoniae at pH 5.0 in 50mM sodium Acetate with DMSO [Streptococcus pneumoniae TIGR4],4XJ9_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with Optactin at pH 5.0 in 50mM Sodium Acetate with DMSO as the cryoprotectant [Streptococcus pneumoniae TIGR4],4XJA_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 5-acetamido-2,3-difluoro-3-hydroxy-6-[1,2,3-trihydroxypropyl]oxane-2-carboxylic acid [Streptococcus pneumoniae TIGR4],4XJU_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 4-acetamido-2-fluoro-3-hydroxy-6-[1,2-dihydroxyethyl]-7,8-dioxabicyclo[3.2.1]octane-1-carboxylic acid [Streptococcus pneumoniae TIGR4],4XJW_A Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with Optactin at pH 7.4 in PBS with DMSO as the cryoprotectant [Streptococcus pneumoniae TIGR4],4XJZ_A Crystal structure of apo NanB sialidase from streptococcus pneumoniae at pH 7.4 in PBS with DMSO [Streptococcus pneumoniae TIGR4] |
| 4XMA_A | 0.0 | 39 | 696 | 1 | 658 | Crystalstructure of the K499G mutant of NanB sialidase from streptococcus pneumoniae in complex with Optactin [Streptococcus pneumoniae TIGR4],4XMI_A Crystal structure of the K499G mutant of NanB sialidase from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4] |
| 4XOG_A | 0.0 | 37 | 696 | 1 | 660 | CRYSTALSTRUCTURE OF THE NANB SIALIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH Optactin and DANA [Streptococcus pneumoniae TIGR4] |
| 4XYX_A | 0.0 | 40 | 697 | 1 | 658 | NanBplus Optactamide [Streptococcus pneumoniae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54727 | 0.0 | 1 | 697 | 1 | 697 | Sialidase B OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=nanB PE=1 SV=2 |
| Q27701 | 3.06e-178 | 71 | 696 | 118 | 756 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| P29767 | 3.88e-50 | 68 | 669 | 220 | 809 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P15698 | 1.62e-06 | 232 | 360 | 41 | 145 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
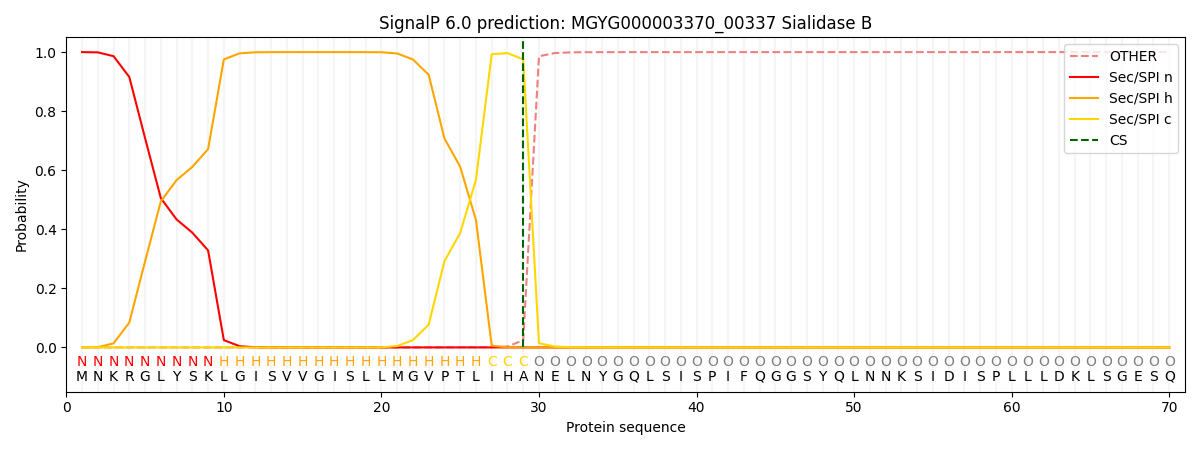
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000310 | 0.998968 | 0.000231 | 0.000166 | 0.000162 | 0.000144 |