You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002724_00458
You are here: Home > Sequence: MGYG000002724_00458
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
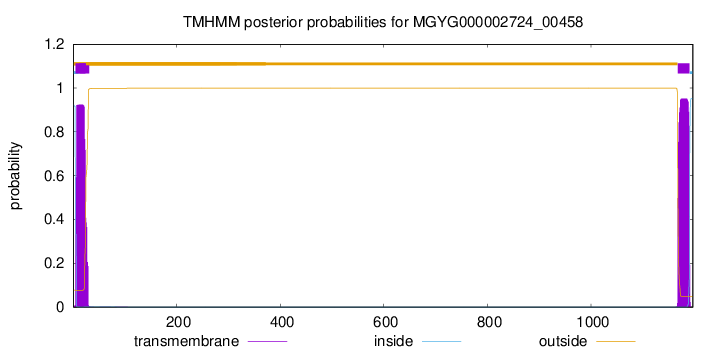
TMHMM annotations
Basic Information help
| Species | CAG-83 sp900549395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-83; CAG-83 sp900549395 | |||||||||||
| CAZyme ID | MGYG000002724_00458 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 70205; End: 73798 Strand: - | |||||||||||
Full Sequence Download help
| MKKILSLFLT LAIVIGLFVP AASAEPTTGA TTTEFAFLVT SDLHGQIYST DYSAGYEASG | 60 |
| TGTTGLTRIA TYIKQMRAKY GENLYLADLG DTIQGAPLTY YYAFNKPEAD DPAMKAFRTL | 120 |
| GYDLWVPGNH EFNYGLDILN RQMDYLTSAA SGKETPAAIM AANYLDAQAR AADPNSWKTW | 180 |
| KNYKPYEIRK FDGVKVAIIG LANPNIPKWD IPANWEGIHF AGIYETYKHY EAEMKAQADM | 240 |
| IAVMTHCGMA YTSEAGDLAM DSVKYLVEKT NSIDFVFSGH EHGTKVLSAN NTDGKSIPIV | 300 |
| QPNTKAKAIG QIIVTYDKDK KAVTSLDATN VSMTQKVSGK TVPCYDIDQE LGQILKPYEE | 360 |
| ATWKDYMLQV IGQSNGDYSA AGLGTAPSAF MDLVNRVQIL GAYDRTGKNT PNNPDDDTPA | 420 |
| QLSISAPLTS GSKENLIPKG DICLGDMFSL YRFENWFYQI RMSGKEVRTW LEYSASKIYE | 480 |
| NNGKLDIKGG LTYYDVIYGD GFSYTIDAAQ PAGSRVVKMT YNAKPVTDDQ EFTVVLNNYR | 540 |
| FNGGGDYVNY LNKHGCNFKA NDPDRIIYSS QFDMINGEDE GQARSLLVSY IKEQTAGQSK | 600 |
| GITPNITSNW KIINSSSAKS DDIVVLYTND VHTYIDNKTG KGLRYSDVAG YRDDLLDTHK | 660 |
| NVLLVDAGDH VQGTAYGGMD SGATILKLMQ TAGYDLATLG NHEFDYGMER ALAIAKNGIV | 720 |
| PYISCNFYHE KDGVKGANVL DSYKIFDLDG KKVAFVGITT PESFTKSTPK YFQDSNGNYI | 780 |
| YGIAGGVDGQ ALYDSVQAAI DAVKAAGADY VIGLAHLGVD ESSRPWTSKD VIAHVTGLDA | 840 |
| LLDGHSHSVV EQELVKDKAG HDVVLSQTGS YLKNLGQLTI TANGQITTKL LSSVSTVQNW | 900 |
| AVKELEDAWI AQIDGELGQK IGSIDSVLGN HNKDGKRLVR SRETNTGDFA ADALYHLFLD | 960 |
| QGVDVAIMNG GGVRNNEITG ELTYKKMKEI HTFGNVACLQ RVTGQMLLDA LEWGARNADA | 1020 |
| DIAKENGGFL QVSGLKYEIH TYIASTVQKD EKGVWTGGPT GEYRVKNVQI FNRATNTWEA | 1080 |
| LDLTKEYKLA GYNYTLRDLG DGFAMFKGAV NELDYVMQDY MVLANYVQSF ENGHVTGYKN | 1140 |
| IDGGDGRITF VFQKAPGTDS GPQTGDETAI GSYVVTSTVT LLALGACLAL RRRKTQK | 1197 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 2.37e-134 | 2 | 1152 | 7 | 1147 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 5.24e-69 | 5 | 547 | 2 | 503 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| COG0737 | UshA | 4.54e-64 | 622 | 1132 | 26 | 500 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09420 | cpdB | 9.51e-64 | 1 | 623 | 1 | 608 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| TIGR01390 | CycNucDiestase | 3.94e-62 | 38 | 623 | 6 | 585 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase. 2',3'-cyclic-nucleotide 2'-phosphodiesterase is a bifunctional enzyme localized to the periplasm of Gram-negative bacteria. 2',3'-cyclic-nucleotide 2'-phosphodiesters are intermediates formed during the hydrolysis of RNA by the ribonuclease I, which is also found to the periplasm, and other enzymes of the RNAse T2 family. Bacteria are unable to transport 2',3'-cyclic-nucleotides into the cytoplasm. 2',3'-cyclic-nucleotide 2'-phosphodiesterase contains 2 active sites which catalyze the reactions that convert the 2',3'-cyclic-nucleotide into a 3'-nucleotide, which is then converted into nucleic acid and phosphate. Both final products can be transported into the cytoplasm. Thus, it has been suggested that 2',3'-cyclic-nucleotide 2'-phosphodiesterase has a 'scavenging' function. Experimental evidence indicates that 2',3'-cyclic-nucleotide 2'-phosphodiesterase enables Yersinia enterocolitica O:8 to grow on 2'3'-cAMP as a sole source of carbon and energy (). [Purines, pyrimidines, nucleosides, and nucleotides, Other] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASN98642.1 | 4.03e-118 | 618 | 1149 | 37 | 612 |
| QRP42178.1 | 4.03e-118 | 618 | 1149 | 37 | 612 |
| QJU22404.1 | 4.18e-116 | 618 | 1149 | 37 | 612 |
| QIX91987.1 | 8.39e-113 | 618 | 1149 | 37 | 612 |
| SET94084.1 | 1.97e-111 | 615 | 1149 | 29 | 607 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3QFK_A | 8.77e-34 | 36 | 551 | 21 | 489 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4Q7F_A | 8.77e-34 | 36 | 551 | 21 | 489 | ChainA, 5' nucleotidase family protein [Staphylococcus aureus subsp. aureus COL] |
| 2Z1A_A | 2.19e-32 | 623 | 1109 | 30 | 495 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 5H7W_A | 5.89e-27 | 620 | 1128 | 2 | 509 | Crystalstructure of 5'-nucleotidase from venom of Naja atra [Naja atra],5H7W_B Crystal structure of 5'-nucleotidase from venom of Naja atra [Naja atra] |
| 3ZTV_A | 1.57e-26 | 619 | 1115 | 9 | 536 | Structureof Haemophilus influenzae NAD nucleotidase (NadN) [Haemophilus influenzae Rd KW20],3ZU0_A Structure of Haemophilus influenzae NAD nucleotidase (NadN) [Haemophilus influenzae Rd KW20],3ZU0_B Structure of Haemophilus influenzae NAD nucleotidase (NadN) [Haemophilus influenzae Rd KW20] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 4.27e-79 | 20 | 1173 | 31 | 1198 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| P08331 | 1.54e-41 | 30 | 623 | 19 | 606 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Escherichia coli (strain K12) OX=83333 GN=cpdB PE=1 SV=2 |
| P44764 | 5.54e-41 | 29 | 613 | 28 | 606 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=cpdB PE=3 SV=1 |
| P26265 | 1.58e-40 | 30 | 623 | 19 | 606 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=cpdB PE=3 SV=2 |
| P53052 | 3.05e-39 | 22 | 623 | 16 | 611 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Yersinia enterocolitica OX=630 GN=cpdB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000310 | 0.998902 | 0.000215 | 0.000194 | 0.000190 | 0.000168 |