You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004508_00761
You are here: Home > Sequence: MGYG000004508_00761
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS2068 sp900769635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RF39; UBA660; UMGS2068; UMGS2068 sp900769635 | |||||||||||
| CAZyme ID | MGYG000004508_00761 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Cell division suppressor protein YneA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1170; End: 2189 Strand: - | |||||||||||
Full Sequence Download help
| MKVIIDPARG GNDTGVSGGG ITEKDWNLAI SKYIYNILKN KGIDVYLTRT EDVDIPEAER | 60 |
| ATKIKNYATT SNDIIVISNR LNEGKDDGAE IIYPLRSTNN LAKALATALE STGQNVNKYY | 120 |
| QRRLPSDLAS DYDYIIRNTS PYETIIIKYG YVDSPEDDIT QLKNYQSLGN AIANTIISRA | 180 |
| SSSGTYTVKK GDTLYSIAKK YNTTVNNIKS LNNLSSNTLR IGQKLKIPLK ETTTPSTTPT | 240 |
| PNTLTYTIKK GDTLYGIATK YSTTISAIKS LNNLDTNTLT IGKTLKIPTT TSYTTHTIKK | 300 |
| GDTLYSIAKK YNTTVNNLIN LNNLKTTTLS IGQTLKVPL | 339 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM50 | 186 | 228 | 1.4e-18 | 0.975 |
| CBM50 | 296 | 338 | 7.4e-16 | 0.975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02696 | MurNAc-LAA | 2.28e-26 | 2 | 177 | 1 | 172 | N-acetylmuramoyl-L-alanine amidase or MurNAc-LAA (also known as peptidoglycan aminohydrolase, NAMLA amidase, NAMLAA, Amidase 3, and peptidoglycan amidase; EC 3.5.1.28) is an autolysin that hydrolyzes the amide bond between N-acetylmuramoyl and L-amino acids in certain cell wall glycopeptides. These proteins are Zn-dependent peptidases with highly conserved residues involved in cation co-ordination. MurNAc-LAA in this family is one of several peptidoglycan hydrolases (PGHs) found in bacterial and bacteriophage or prophage genomes that are involved in the degradation of the peptidoglycan. In Escherichia coli, there are five MurNAc-LAAs present: AmiA, AmiB, AmiC and AmiD that are periplasmic, and AmpD that is cytoplasmic. Three of these (AmiA, AmiB and AmiC) belong to this family, the other two (AmiD and AmpD) do not. E. coli AmiA, AmiB and AmiC play an important role in cleaving the septum to release daughter cells after cell division. In general, bacterial MurNAc-LAAs are members of the bacterial autolytic system and carry a signal peptide in their N-termini that allows their transport across the cytoplasmic membrane. However, the bacteriophage MurNAc-LAAs are endolysins since these phage-encoded enzymes break down bacterial peptidoglycan at the terminal stage of the phage reproduction cycle. As opposed to autolysins, almost all endolysins have no signal peptides and their translocation through the cytoplasmic membrane is thought to proceed with the help of phage-encoded holin proteins. The amidase catalytic module is fused to another functional module (cell wall binding module or CWBM) either at the N- or C-terminus, which is responsible for high affinity binding of the protein to the cell wall. |
| pfam01520 | Amidase_3 | 3.56e-24 | 3 | 176 | 1 | 172 | N-acetylmuramoyl-L-alanine amidase. This enzyme domain cleaves the amide bond between N-acetylmuramoyl and L-amino acids in bacterial cell walls. |
| PRK06347 | PRK06347 | 1.00e-21 | 178 | 337 | 400 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 4.05e-21 | 164 | 337 | 311 | 523 | 1,4-beta-N-acetylmuramoylhydrolase. |
| COG0860 | AmiC | 2.98e-20 | 1 | 178 | 43 | 226 | N-acetylmuramoyl-L-alanine amidase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVK20078.1 | 8.64e-39 | 3 | 288 | 2 | 284 |
| QIC05530.1 | 8.96e-26 | 3 | 172 | 85 | 248 |
| AGY48195.1 | 6.59e-25 | 2 | 287 | 3 | 302 |
| AGE61023.1 | 2.42e-24 | 2 | 287 | 3 | 302 |
| AYB39952.1 | 2.58e-24 | 3 | 172 | 157 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EMI_A | 1.03e-08 | 1 | 177 | 5 | 175 | ChainA, Cell wall hydrolase/autolysin [Nostoc punctiforme PCC 73102] |
| 4UZ2_A | 6.52e-08 | 185 | 233 | 4 | 52 | Crystalstructure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ2_D Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4UZ3_A Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_B Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8],4UZ3_C Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose [Thermus thermophilus HB8] |
| 4XCM_A | 5.93e-07 | 185 | 233 | 4 | 52 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54421 | 1.01e-16 | 172 | 292 | 14 | 134 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| Q49UX4 | 8.08e-16 | 170 | 287 | 13 | 129 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| O31852 | 5.28e-14 | 172 | 292 | 15 | 136 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| Q8CMN2 | 3.46e-13 | 178 | 288 | 75 | 191 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=sle1 PE=3 SV=1 |
| Q5HRU2 | 3.46e-13 | 178 | 288 | 75 | 191 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
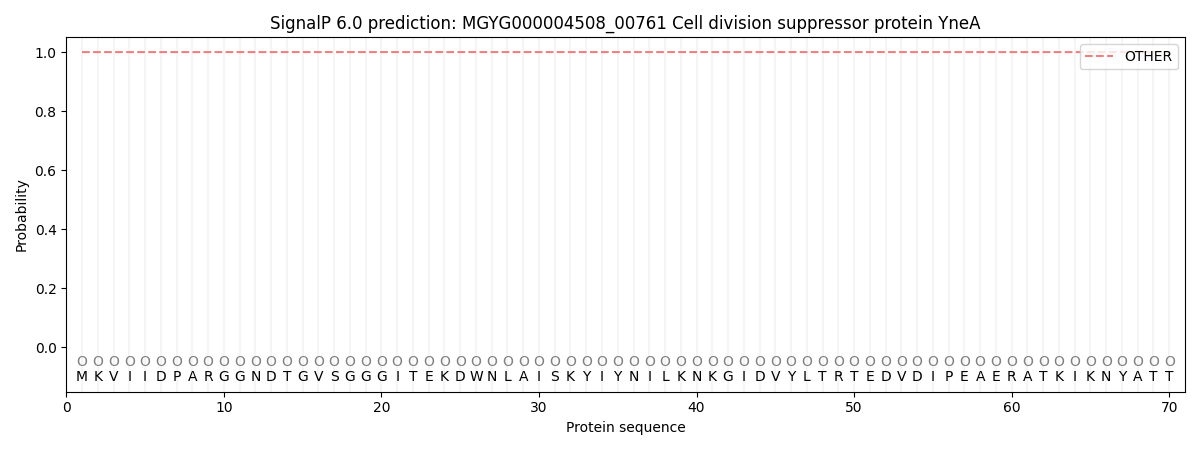
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000061 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
