You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000919_00171
You are here: Home > Sequence: MGYG000000919_00171
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Niameybacter sp900551325 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Cellulosilyticaceae; Niameybacter; Niameybacter sp900551325 | |||||||||||
| CAZyme ID | MGYG000000919_00171 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 188890; End: 195201 Strand: - | |||||||||||
Full Sequence Download help
| MNKKLIASLL SVSMAAGAGV QTPVQLMAST VPVTKLSTTN VFNEQVIETQ NITSSVFSPI | 60 |
| KNVHYTEYEK AFKVTGIKTY TNNGGQYGSS ALKYAFDGDI GTHWETGKQN TNEYKNEVTI | 120 |
| TFDNIETINR IAYAPRPGNK GHVQKMSIYA SLTEEGEDFQ LVATGQTEVR SNVSEFRFQN | 180 |
| TQFKRLRFVF EQANQEWAAA GEFMFFKEDA VADKMATLFT DGTHSQVSEA FRSSEQLDAL | 240 |
| EKEAQAHPFY ETEYKENIVN ARRLLENKEV QYTEAQVTKL TLSGDLLEAY DEVYRLQADG | 300 |
| IKGITNNGGS YAENAIERAI DGRLDTKWHS NVKNSESFTN EVVIELNELT TLNRLVYAAP | 360 |
| RGSNRGFPEA FEIYASRTSK GNTFELVSTG KAQVTQDLIE IKFNPTEFKR LKFVFKKGYE | 420 |
| DWACSSEIRL YREDTVQDAM EKLFTNGLMM EVSSEYNSID TINKLEEQVN SHPLKESYLE | 480 |
| DIKLAKEIVY NKDLFKDTRV VTGEQRGRYK EESEKRSING YAYTSLESIG KYVTPGEEIV | 540 |
| VYVDADATGV LPKLCFGQVG RGKGDWRRWA DLKPGRNVIT APTNINPSAL YLFNDAKPSD | 600 |
| QAYAPRVRVE GGTDFPLYIH GETDPQAFFE EVKAYAQKVE YSDDAFVNGN PEGKVFNIAE | 660 |
| FVSKNIVITT SAKGAVAGMT EAYSKGYDMA DTMDGWEELY DMFQTFMGFE KNATEEKHSY | 720 |
| FPNKFISRAF QGVPLGFADH GYTGYLGSDN AERDGGFFKL LAMPPQMPGN DNWAYGHEFG | 780 |
| HIFNTKYMVH GEVINNLYPQ EYRRIKGIDG DRCNWNGLMK RFQGETVNLG FWENLGILSQ | 840 |
| LNIAYGYDTY AKVSRAVRDN TELIASIQGS ELRRLAVAYS LGVGENLLDF FKGWNYTDVT | 900 |
| PEMEEAVKHL PKPTRKIEYL HSGAYDYKGN GFSEDIAIKV TSKLDTIAKT NKLSFEIDEN | 960 |
| NQKDLLGYEI LKDGKVIGFT KDNSFTVKNA DIQASENYEV VAYAKDLSIA KPVSVNSFAP | 1020 |
| TLKVQQEKVT LKLGETFNPV DYIKGSNYKG DNITAQVNIT HNVDTTNKGT YEVKYTLLDQ | 1080 |
| GVTVEKVVQV EVVSDYEYLS DYQWQSVETE WSTPRRNNNI TSTVNGEVKV FEKGFGIHAN | 1140 |
| GKIVYDLSNM KHDRFEALVG IDRSIPEQAN SSVIFKIIGD GQVLATTKVM RYGEDMVYID | 1200 |
| VPIKGVSELV IEIDDANNSI SSDHAIIANP KLITNNVLPT LDVEELTYIK LNSQFDTMAD | 1260 |
| VVANDVEDGN ILEKVQVETN GFNTGKTGKY NLSYTVADSD GNTVTKERTI VVYSEVDYLS | 1320 |
| DRNWENARTD YGSVNKDKSS SGASIKLGVK GETITFAKGV GTHANSEIVY DLSKLSGSVF | 1380 |
| FETYVGVDRN IEEQNRSSII FKILGDGKEL YNSGLMKRSD DAKHVKLDLT GISKLELIVD | 1440 |
| KNGSDNAFDH ANFGDAKFLV LDAVPTLEIP KDEAVKVGEH LEDIYGTYSA TDAEDGDLTS | 1500 |
| KVEVMGTVNF KQPGHYTITY RVTDNDGNEV IKTRTIAVVD MEDFTYLSDY NWASATCGWG | 1560 |
| SIKKDLSPSD NIIRLTDAKG QEVQFEKGLG THARSVLKYD LTNKEAEYFS AYVGVDRAMY | 1620 |
| NSIASVSFEV WLDGEKVTET GVIYSKDPME YLEVNVAGAK ELMLVATDGD NGNGSDHAVW | 1680 |
| GDAKFHYANE NNIKVDTEAL VQLIEEVNAL NGENYTKESW NKLVQVRDEA VALLEGEKTQ | 1740 |
| VQVDEMVNKL VEAKNALVVV GNKQVLESLV ASVNDLEAHI YTQVSWEALL AQVESAKEVL | 1800 |
| TIEELTEQEI NEAAQKLQDA IDRLEVSKGK VGLYELLKVA NTMKQSDYKE GADWETFAEV | 1860 |
| TAEYNKIYWA PTVHSELVFE IVLDYYSSLI QSIEAHRIPE NHKGELEAVV ANVEDLEAHI | 1920 |
| YTQLSWESVQ KNLVEAKKVL GLEEVTQEVI DEVAQNLQDS IDSLEVSKGK AGLYELLQIA | 1980 |
| NAMEKSNYSK EADWATFEEM TAYYNELYWA PTVHSESMYE LVLEYYNGLI QSIEAHRVSN | 2040 |
| NNIDQVEENQ VDGDALEVEQ PQINQPEAGQ VEESQVNESQ LEEGQLEEGQ QVMDQTQVEE | 2100 |
| LER | 2103 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1545 | 1685 | 2e-42 | 0.9925373134328358 |
| CBM51 | 1318 | 1458 | 1.4e-39 | 0.9850746268656716 |
| CBM51 | 1098 | 1232 | 4.7e-34 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 9.77e-50 | 1545 | 1686 | 3 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 2.07e-45 | 1318 | 1458 | 4 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.68e-42 | 1095 | 1233 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.29e-40 | 1545 | 1686 | 5 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| smart00776 | NPCBM | 1.35e-38 | 1318 | 1458 | 6 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35289.1 | 0.0 | 55 | 1757 | 275 | 1966 |
| SQG38452.1 | 0.0 | 55 | 1757 | 275 | 1969 |
| AXH52096.1 | 0.0 | 55 | 1735 | 275 | 1946 |
| QPQ34479.1 | 0.0 | 68 | 1719 | 63 | 1715 |
| AHN23111.1 | 0.0 | 68 | 1766 | 63 | 1763 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JFS_A | 8.72e-155 | 68 | 1016 | 43 | 981 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 5.35e-123 | 504 | 1458 | 28 | 975 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JND_A | 4.33e-100 | 68 | 453 | 61 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 3.21e-98 | 68 | 453 | 61 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 5KDJ_A | 1.86e-65 | 434 | 1079 | 24 | 656 | ZmpBmetallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124],5KDJ_B ZmpB metallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 2.37e-10 | 1318 | 1472 | 1164 | 1323 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| E8MGH9 | 1.22e-07 | 1631 | 1868 | 1591 | 1851 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
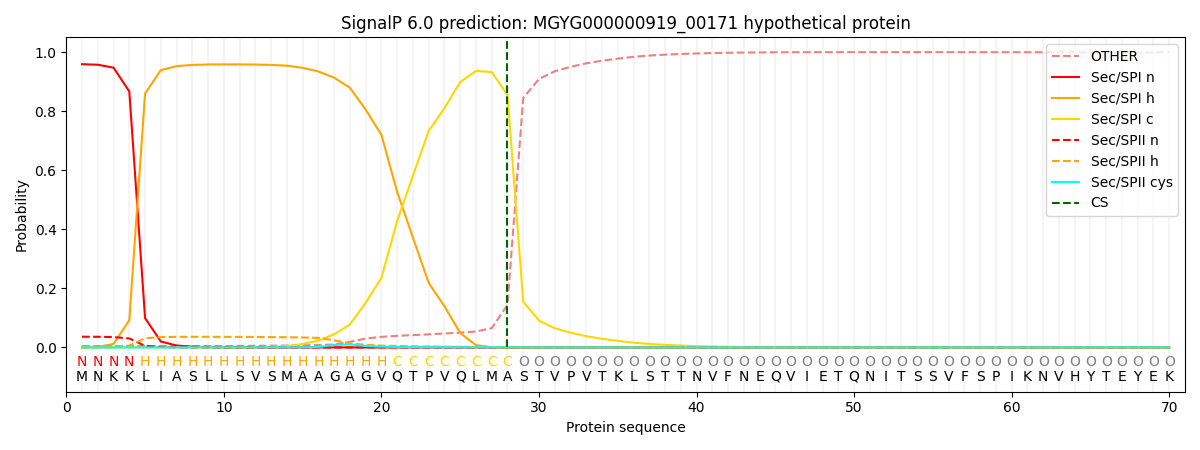
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.008119 | 0.951712 | 0.038620 | 0.000850 | 0.000377 | 0.000290 |
