You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001018_00230
You are here: Home > Sequence: MGYG000001018_00230
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
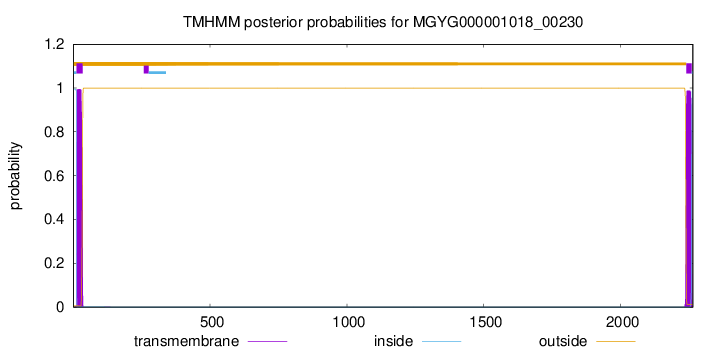
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900751785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900751785 | |||||||||||
| CAZyme ID | MGYG000001018_00230 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14220; End: 21014 Strand: + | |||||||||||
Full Sequence Download help
| MKKRQRRRGR KILCGVLAGV TVSSMAAVPG LASTDWPDMG PANALTEDYM GLHRSVRDSN | 60 |
| IRPDDILTLD SDNDLLKRAF SRSVDYAYDE LVVGPKQFPL DEQNEQRISK HYNGEGYYTS | 120 |
| TAIPAIWGTY HGLPGQSYRE CFCARDVAHT SEAGALLALN EENYQMLSRF AEESADLYWR | 180 |
| DVFHSEDGGW STKSANQKWW TLWSFDFYGD AYYMDAGFQE LPAPFEVMDK IWTQYRWTGD | 240 |
| ERWIDDTMIN YGMQLHSYDK FMKNHDLDGD GLADNGKNGS ILPTLYEFEA SGYLQESDVD | 300 |
| WEKMDEPTGD PSYPYESTLV IKPDFFMNHS FYELPLTMKL IGKSGNTDVE QGENAYIVRI | 360 |
| RNKEYDAEKP AVLKTFEQMS SRNNYADIHD VEIPVYIPEG VTFEVYNSAE KMNEEYYTVS | 420 |
| DNGDGTKTIT VKEDFFRHEY ADNGIYRTNT GIVVRFSDGS SGMAIIKLDD SNVNGDLNVV | 480 |
| SIKKNGTDVT DSLEFDPSIN TTYLDYDAAE LADTVYTLTI RHSDPVTPKK EQAGSLQEYC | 540 |
| RIASDLTRVT TAGDTAGTQY QALLAMAGMI RARIAAGDTT FQPFHLEKGV KVNEGEPITL | 600 |
| TEEMAQEYED RAAGIRDIIE SEWRDESAPQ IYARAVNKLG EKDMGWGHEN SFFMPMKELL | 660 |
| NPGEDTIKYL NFIQKNALLE DQLNEEAITY LPESFYTHGQ NVAGWQWTKV GLDRLDPGKK | 720 |
| TETEILRTYP EIALTNISNV VNLMMGVEPY AADNEVTTLP RLTDEVGWVT VNNIPLGQEG | 780 |
| REALNNDGRD SDGNGRPTGY GKPMVNNSFD LTHVGTAQST LTNTGDETIT WNAEFYGTYD | 840 |
| KLYKTVDGEV TELTEGIYHK DTAPASTYCT IKVEPGETIT VGTQKDPNGA KPNTDGMIYL | 900 |
| SDQEAAFEDY PDDKPVQKDQ TIGGTALYNY TFTEHYDKGL GVKPNTTIRY YLPEGTEAFK | 960 |
| AVPTITDFVR GQDPQETAAE ITFRVYLDGE LKQTVTLNNE KPTGDELDIA IDGQSVLDLV | 1020 |
| AVSENQNIQE SAAWLDARIL VEGQDEIPSD LPDNKLYLTS MQAVDESGEV SADQSTQNAA | 1080 |
| ALEIDGHAFG RGIGSNGSSE VTYELSGLYE KLDAVIGIQD GNSGSAVFKV YLDDSQTPMF | 1140 |
| EHTADADSGE EKVSIDLSVD GKRADRIRIV TEGDTHSVWG DMILTIGDDV DLTDPAIIAK | 1200 |
| RIASAEDIGL GQKEYNLPEI PVKNADDYTI EIASSSEEAV LALDGTITLD DNEHDIAVSF | 1260 |
| RITEKEGQKR SAVTREFVIY APGKDKISLT KLKWTDSVSE NAPVTINEDI KGNILKLDDT | 1320 |
| EYSRGIGMPA APASQITYEL EDGLYKRFTA TVGAAQNSSV NDNDLFRVFV YAVDQNGKRT | 1380 |
| ELYASPVFVK GPVDQNNHLN CDLCKAEIEK YGLAAQDTVD VDIPSGYRQL VIRTERIGDS | 1440 |
| FWGSCAIADP YFYLNSPYEG KEIESFEPVS VQTVKGVKPV LPDMVKAIYK DMDVPRNIRV | 1500 |
| TWDEIDESLY QEVGQFTVSG TVEGSDKRPE CLVEVKDKII LTAENASVVT TIGNAPVMPE | 1560 |
| TVRVQYNDNS SGEEKVTWDP MDESNWAQAG KFTVMGTVDG TDIRVGCIVR VDDPNKVIVA | 1620 |
| DMEPESETAM ESVQRGTFRI NGKDYENSIF AHADSELIYV LDGQYTRFTT TVGIKTEGDN | 1680 |
| PNVIFRGYGI DENGEETLLY DTNPVTRVAD PSKPPQTTDE NGYYYVGGNV SYDVDVDVTG | 1740 |
| FQKLKLVADV VGNPNTDHAT YGTPILYKEG SEFGKNFHTI TVTADEGGAI WPNGRVVVPD | 1800 |
| GESKEFIITP NPANPVLPEV KEMAINEFKL DETEQTVTDN RFVLENVDQD HTISVNFRDA | 1860 |
| SEVPDVKYNV QVVNGSGSGE WAAGSTVSIT ADAAPEGQVF DCWVSDDGVE FADAQAASTT | 1920 |
| FVMPEKDVTV TATYKESQVS KKTLEYFLNS AKEHLANGDV DDAVESVQKL FEEAIAEGEA | 1980 |
| VMAKEDATRE EIMNATAKLM KAIQALDMKA GDKTDLQMAV ELADMIDLSK YVEAGQKEFT | 2040 |
| DAFAAAKDVL ADGDAMQADI DSAWSALVDA LDNLRLKANK DALEDLLDEV SGLDLSQYTE | 2100 |
| ESAAVFEQAL MRANEVMADE TLSVEDQAAV DEAVQALQSA KDGLVLKSDD TGDGDQNTGD | 2160 |
| GDQNTGDGDQ NTGDSDQNTG NGDQNTGDGS QNNGGSQDTG NGGKDTGSPA DGQNGKAPAD | 2220 |
| GAVQKTESGR KAVKTGDTAA AGMLLAVLLI SGVTAVTVLK KKRA | 2264 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1061 | 1181 | 2.3e-17 | 0.8656716417910447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.57e-19 | 1621 | 1766 | 7 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.96e-15 | 1057 | 1185 | 4 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 2.83e-14 | 1634 | 1766 | 35 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam18998 | Flg_new_2 | 4.35e-13 | 1868 | 1935 | 1 | 71 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| smart00776 | NPCBM | 8.99e-13 | 1058 | 1185 | 10 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYJ80995.1 | 2.05e-142 | 30 | 1184 | 16 | 1064 |
| QYJ92352.1 | 1.78e-141 | 30 | 1184 | 16 | 1064 |
| QUH30699.1 | 2.42e-139 | 67 | 1399 | 37 | 1009 |
| ABO24155.1 | 1.15e-136 | 30 | 1184 | 16 | 1064 |
| AQM60758.1 | 3.43e-52 | 1461 | 2150 | 1360 | 1999 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NIT_A | 1.68e-06 | 1553 | 1605 | 902 | 954 | ChainA, Beta-galactosidase [Bifidobacterium bifidum],7NIT_B Chain B, Beta-galactosidase [Bifidobacterium bifidum],7NIT_C Chain C, Beta-galactosidase [Bifidobacterium bifidum],7NIT_D Chain D, Beta-galactosidase [Bifidobacterium bifidum],7NIT_E Chain E, Beta-galactosidase [Bifidobacterium bifidum],7NIT_F Chain F, Beta-galactosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.84e-11 | 1462 | 2067 | 1298 | 1868 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
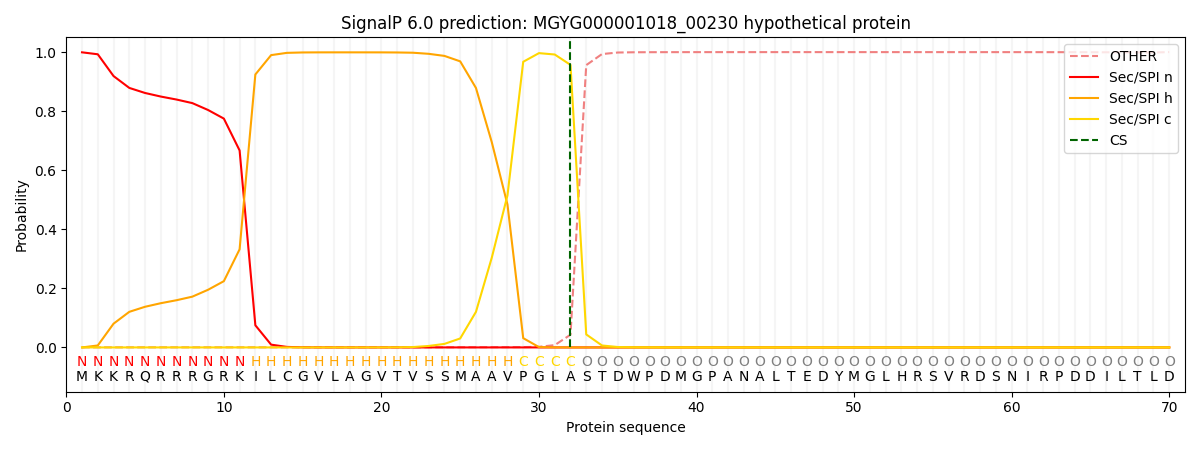
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002109 | 0.995971 | 0.000556 | 0.000771 | 0.000313 | 0.000240 |