You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003140_00276
You are here: Home > Sequence: MGYG000003140_00276
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
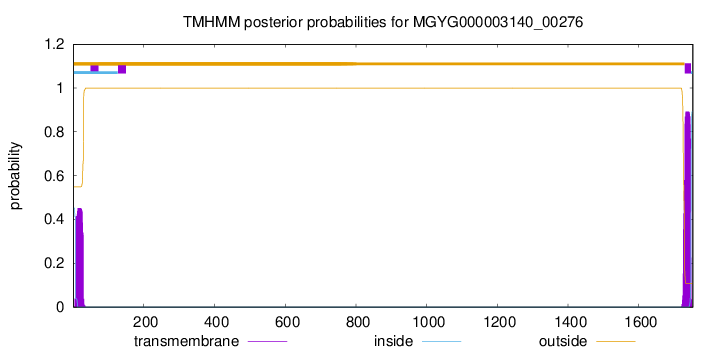
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp000758755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp000758755 | |||||||||||
| CAZyme ID | MGYG000003140_00276 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21519; End: 26780 Strand: - | |||||||||||
Full Sequence Download help
| MIDKRVFWRP LAILGALALS LPGSAVAHAT EGATDTPVIL NAPTTISAGQ LRATVSHEFP | 60 |
| QVTGYTFAGN PVGGRAETLH QVTIDGNQYT VDTVEAQRVS DQKVAYTVTF VGSEITMKAE | 120 |
| IEVQEITSHA QGTEGAKRPT LTFRITEMTG AHTVEIPGHG LVSVNADQGG AYACGITGVS | 180 |
| RGAANGDSYA GVDDTFASIT ASTPIDYYEQ PSAYLMVNTN KVAVGMETNA TYDEPHGYET | 240 |
| NWNSRWKRQV IEQNGIKTLI AQNGQWTYRS EAATDAIGDE ERPYTTLVFT GDANSSGSVD | 300 |
| WQDAAVAYAD ITPEITGAAD NHKWVVTHIP FDFGSAATHP FLQIADDVKR VSLATDGLGQ | 360 |
| RVMVKGYASE GHDSGHMDYG GNINTRAGGE ADFGTLFTST KDVNAIYGVH VNTTEAYPEA | 420 |
| NSFRSLPFTG GRGWNWLNQS YYVNQRDDLG NGGAVNRFQE LRNQFPLSKY PNFRWIYIDV | 480 |
| YYGSGWQADR LGNELNKMGW EVGSEWADRF ERHSLWSHWS NDEHYGGATN KGLNSQVIRF | 540 |
| VDNANKDNWN PNVVLGYPQI VEFEGWTGHQ DQDAFYRNIW SNNLPSKFLQ NSRIMRYDTA | 600 |
| DAGDGKTKHT YTFANGTVAS GSTAVTAQTE ASYVVGTIQA DMSSSREFVY DGATVLRGDS | 660 |
| YLLPWKDNGT SAGADRLYYY NPAGTATEWT LTKSYQGVGS LALYRLTDQG RVKVTDLAVS | 720 |
| GGKITIPASV YDGVAEGDRD HTAFVLVPEN APAAVSTPAW GAGTVFADPG FNSGSLSGYT | 780 |
| TTGSVSVVKD PHGDPVAQLG AGKASLSQEM TLNAGTYQGS AWVQINDGKT RKVTISASGD | 840 |
| GVTSAGSQQR NGDLGATRDK PVTTVFEDFE NIAEGWGPFV KGDAGNITDP RTVLASLNAP | 900 |
| YTQRGGVLPD RSRKATDDAI AGHWSLKSHE ENGGLVYRTV PQTVNLQAGH RYRISYDYDN | 960 |
| AVSGSYYWVT AYDSAGADGV SANYLDSTLL PQTERPSHFV EELNVGACGS YWVGLYNVQG | 1020 |
| GKSGNDFSID NFRVEDLGTT ADVPACAAAS IATQGSAIEG KTVQVTTTLV NNESADVTNV | 1080 |
| RSELSLPAGW SAQATSPTTA ETLAKGETFT TTWTVSIPEG AGGNAVTLTH QAHYRIGDED | 1140 |
| RTAVGTTSVT ALGSIRSNAV NYLSDIPFSS TWNSNGWGPV ERDSEVGEDQ AGDGDPISMN | 1200 |
| DQPYAKGLGA HAPSDLGFEL DGKCTTFKAT VGVQDGQWSG SVTFHVIGST DGSSWSDVVP | 1260 |
| ETSVIRGGSE PRDISGNVRG MKYIRLIVGD GGNGNGNDHA NWGNARVLCG NATEGGDTPA | 1320 |
| PEPTPTLTLK DYTGALQLVS APAAYEANPA SNMFDKDLTT FYDKDWVNLT DHPSTIDLAL | 1380 |
| FEGSDPTGAQ PVSIGGLSIA GRAGQANGRP KAYEVYVGQD AANINQKVAE GTLRNTALAQ | 1440 |
| RITFDEAVNA KYVRIRVLSN YKTKADQPDG LLAIAELGVL VPDGTTRAAV SGSDRSAGTR | 1500 |
| SARRARSLDD EQALTVNTNA EGLVGQNYLD VQTVNATDPA ANPETVIDRT WAENWMAADQ | 1560 |
| KHRTNSDPSS FQRVPVTFTV AKDGTKVRFT IAADEGDAPV LFDNLRMVKI ERPATNDLLG | 1620 |
| VDCMADPTPE ACTTMRANAR IGLSTALSVS LPAPTPDPTA DPSAQPTTDP TTQPSGDPSP | 1680 |
| QPSADPSQSG SQQASPATQE QAGAASAKDG KKKSTRKSKK SSLARTGGET SAILISAALL | 1740 |
| TAAGYALRRR RMN | 1753 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 50 | 728 | 2.4e-165 | 0.9801980198019802 |
| CBM51 | 1162 | 1306 | 1.8e-34 | 0.9776119402985075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12905 | Glyco_hydro_101 | 3.27e-127 | 302 | 576 | 1 | 273 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| cd14244 | GH_101_like | 5.43e-117 | 317 | 599 | 3 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam18080 | Gal_mutarotas_3 | 2.66e-50 | 46 | 301 | 1 | 243 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| pfam08305 | NPCBM | 8.54e-39 | 1160 | 1307 | 2 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 4.53e-30 | 1161 | 1307 | 5 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEG53803.1 | 0.0 | 9 | 1645 | 8 | 1630 |
| QQM68206.1 | 0.0 | 7 | 1645 | 6 | 1635 |
| QQO77976.1 | 0.0 | 9 | 1645 | 14 | 1640 |
| AVM61293.1 | 0.0 | 9 | 1645 | 14 | 1640 |
| QDW61529.1 | 9.15e-302 | 44 | 1346 | 73 | 1371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6QEP_A | 2.21e-105 | 45 | 1037 | 11 | 1116 | EngBFDARPin Fusion 4b H14 [Bifidobacterium longum] |
| 6SH9_B | 2.29e-105 | 45 | 1037 | 11 | 1116 | EngBFDARPin Fusion 4b D12 [Bifidobacterium longum subsp. longum JCM 1217] |
| 6QFK_A | 2.29e-105 | 45 | 1037 | 11 | 1116 | EngBFDARPin Fusion 4b G10 [Bifidobacterium longum] |
| 6QEV_B | 2.29e-105 | 45 | 1037 | 11 | 1116 | EngBFDARPin Fusion 4b B6 [Bifidobacterium longum] |
| 2ZXQ_A | 2.87e-105 | 45 | 1037 | 26 | 1131 | Crystalstructure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 5.32e-240 | 9 | 1341 | 13 | 1330 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q8DR60 | 3.67e-97 | 45 | 1040 | 333 | 1421 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
| Q2MGH6 | 4.86e-97 | 45 | 1040 | 333 | 1421 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
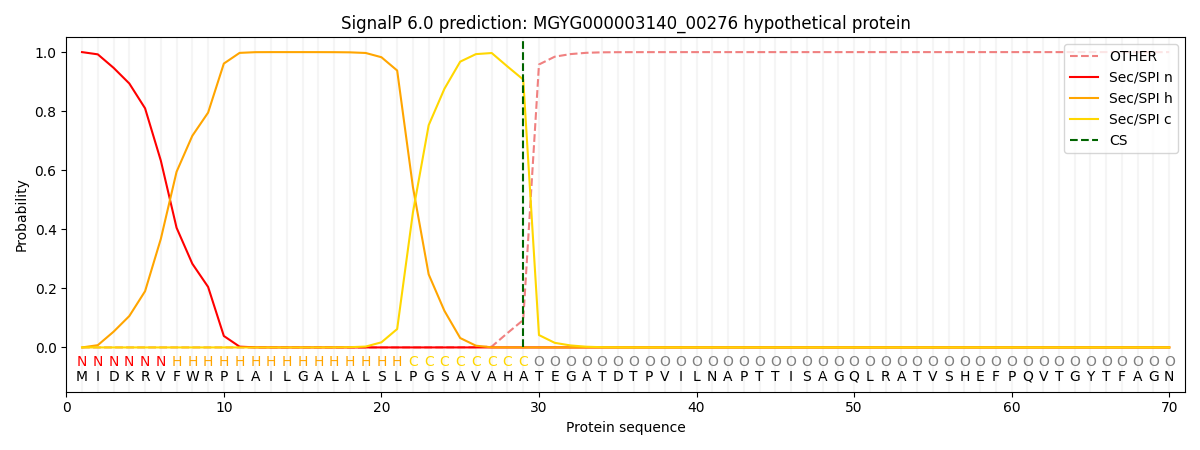
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000422 | 0.998661 | 0.000226 | 0.000291 | 0.000195 | 0.000169 |