You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004594_00277
You are here: Home > Sequence: MGYG000004594_00277
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
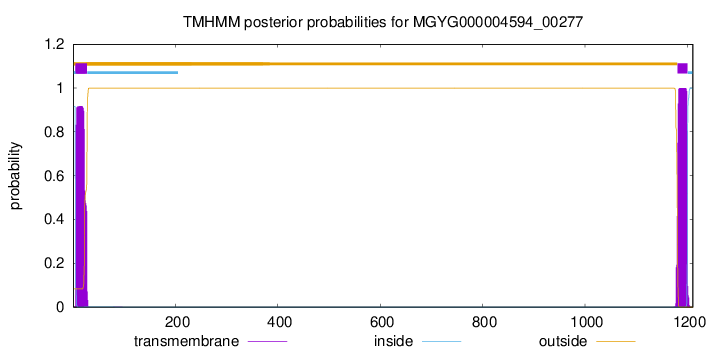
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; UMGS1663; | |||||||||||
| CAZyme ID | MGYG000004594_00277 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47383; End: 51018 Strand: - | |||||||||||
Full Sequence Download help
| MNKKIFSFVV IALTFIAFNL YNNEIFAIDN IENENYIYLS DVSYIEDKSF AQGNHTIHLD | 60 |
| KNEDNGMITL NVDNSSKSFI KGLCAWATSE IVYDLSSYNY DYFTSYIGVD ISEQNSYFNT | 120 |
| GVKFYIYTSN DGENWDKQFE SETLDAWSSA QFVKIDIKNV NYLKLVADDN SDSWWASWYD | 180 |
| EAVYANAKLI KENYEENTSN IDFIKTVDEY DSIIKNHFND DINGDYELTL LQREFVNNVG | 240 |
| YDVLQALAQY KEEYKDTINW LMHDSQTLKL YLVGGKPEGS YLNSIKVLSD LYNTYKDDLL | 300 |
| NTNKTKYGTT LGDLYRTMML SLSLTESGNV YLWVDGTVQS DALTRYEIYK NLHLHEGQDA | 360 |
| ELIENKIFES LTVEEMRWVM NTVIDDEEIL WLNDYVRNEG KGATSPYNYI TYTFDYDYTL | 420 |
| DKYYNSENYD KWNQKYHLSK YNITYKSGHP KLWVVFEEGS VCGGLSKTGS CIWGAYKGLP | 480 |
| NTCVSQPAHC AYIYYTQNDN GNGIWNLGNN VSGWGKSGRT EHLNVRTMND WGNGNYTSGW | 540 |
| NANYILLAQA AQNEYEKYEK AEEVLMLADV YKDDSQKLEK IYRKAIEIED INFDAWLGLV | 600 |
| NLYTNDSTKA ESDYYTLAKE IVKVYTYYPN PMYDLLNLVK PYFTSVEYST SFTLLQTNAL | 660 |
| TAASKATNVE SIQSQAVKQV AEQLLGNVDT TIANFSFDGD NAGKIVLSSR YDGSDVAWDY | 720 |
| SLDGGITWTK TQEHIVQLTN EEIASITSEK DIRVHIIGVD YSNENIFIID IQDSLGLPLT | 780 |
| LYANDLENKL IGATNSMQWK YNDSDEWTSY SVQEPDLTGD KSIIVRAGAT GLYLAGSTSA | 840 |
| TYTFTKDSLP DTNKYIPIKH LSIHYVSSEQ TGSDDAINCI DGNINTIWHT SHNASDSDRT | 900 |
| IIIKLDEPVY LSSVQYVPRQ FGTNGRAKNA ILYVSMDGVN WTIAGNAANW ENNSNPKTIE | 960 |
| LNESTKTQYI KFVTTENWGD GRNFASAAMF NLFEDTTKKI PPTAEIEYTT NNDNTVTAKL | 1020 |
| VNCSTNITIT NNAGSNIYTF TENGQFTFEF IDKYGNVGAA TAVVDWITNS SDKDDNKPGE | 1080 |
| DIDKPSDDNQ KPSGDTNKPN DDNQKPSGDV NKPNNDNQKP SGDTDKPNDD TDKPSDNNSN | 1140 |
| SGENNNNNSN SKPNEDIQNS GKDNTISSGK LPNTGMNYKM PVIIALLTFT TILAGIIAFI | 1200 |
| KKLRYKNRKC L | 1211 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 38 | 189 | 1.9e-23 | 0.9850746268656716 |
| CBM32 | 867 | 988 | 5.6e-18 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.40e-25 | 36 | 190 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 5.22e-18 | 37 | 189 | 5 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 7.36e-18 | 867 | 988 | 5 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.19e-06 | 866 | 977 | 16 | 131 | Substituted updates: Jan 31, 2002 |
| pfam07738 | Sad1_UNC | 0.002 | 892 | 980 | 22 | 115 | Sad1 / UNC-like C-terminal. The C. elegans UNC-84 protein is a nuclear envelope protein that is involved in nuclear anchoring and migration during development. The S. pombe Sad1 protein localizes at the spindle pole body. UNC-84 and and Sad1 share a common C-terminal region, that is often termed the SUN (Sad1 and UNC) domain. In mammals, the SUN domain is present in two proteins, Sun1 and Sun2. The SUN domain of Sun2 has been demonstrated to be in the periplasm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35280.1 | 4.34e-221 | 19 | 998 | 31 | 991 |
| AQW23377.1 | 1.47e-218 | 33 | 998 | 45 | 991 |
| ATD49073.1 | 1.71e-218 | 33 | 998 | 50 | 996 |
| BCL58565.1 | 1.85e-179 | 32 | 1067 | 76 | 1118 |
| QTC11463.1 | 7.62e-137 | 29 | 1000 | 132 | 1101 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 2.28e-12 | 856 | 994 | 11 | 143 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 4LKS_A | 3.45e-11 | 865 | 988 | 30 | 160 | Structureof CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LKS_C Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LQR_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124],4P5Y_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 5.68e-11 | 38 | 189 | 613 | 752 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRM_A | 1.30e-07 | 39 | 326 | 86 | 337 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRL_A | 1.36e-07 | 39 | 326 | 108 | 359 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2MGH6 | 1.88e-11 | 878 | 988 | 1510 | 1620 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 4.18e-09 | 878 | 988 | 1510 | 1620 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
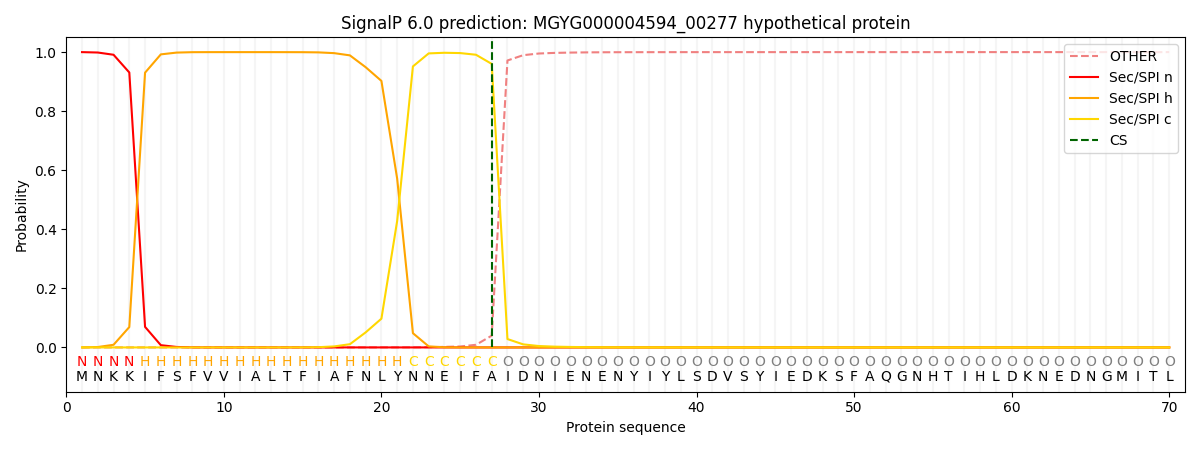
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000982 | 0.998164 | 0.000242 | 0.000196 | 0.000185 | 0.000176 |