You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000220_02570
You are here: Home > Sequence: MGYG000000220_02570
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
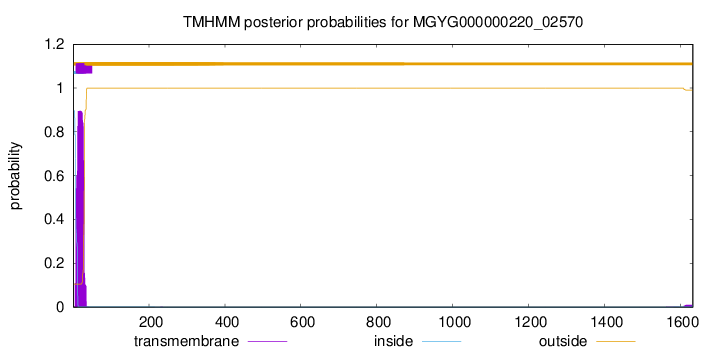
TMHMM annotations
Basic Information help
| Species | AM07-15 sp003477405 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Butyricicoccaceae; AM07-15; AM07-15 sp003477405 | |||||||||||
| CAZyme ID | MGYG000000220_02570 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36670; End: 41571 Strand: + | |||||||||||
Full Sequence Download help
| MRPRNALRRV SALLLTLAML IGLMPQWSGE AQAASYMQPY LDKMVNWGFM RGDIQGNLRP | 60 |
| DNNITRAEFV TIINRAFGYK KIGTTPFKDV KESDWYSDDV AIAYNVGYIN GTSKSTFSPT | 120 |
| KEITREEAAV ILARNLMMQA SVGESTSFTD SRELSSWSRG LVDTAASYKL ISGYPDGSFR | 180 |
| PHNPITRGET AIMVVNAVGT PVSKAEEYQL GSVYGNVLIT ASGATLKNTT IAGNLYVTAG | 240 |
| VDLGNVLLEN VTVLGEIVLS GGGVSEGGDD SLVLRNVNGS KIIVDNLKNQ QVSLRVEGDG | 300 |
| TVDVTSVRTD TYIVDRTASG YGLKEIQLDG EDGRVLKLSG NVKKVVNYTP KSYIGVESGQ | 360 |
| VETITIDEKA TGSTLHIAAG ATVDQLNLDV GTTVTGDGDV ANLTVNADGS TVSMLPDQII | 420 |
| IRPGVEANID GEVMDTEAAA ESSSDPRMLA GYPQITDLAP TSATAKFAAN KKGTIYWAVT | 480 |
| SVTDGSVNAQ DLINPPSYSS TILKKGSVSV TGSGQTATAK ISGLTSDGAY YLSAVLVDAR | 540 |
| EDQSPVKVIS FTTPDNTVPA FATGYPYLSR VTNVSAQVTT MATKTCRLYY AVLPKGSKAP | 600 |
| TGEDFKANAV TGNLGYGSLD VSKNTPYTFD VNNVPLEELQ SYDLYLWLTD IEGGSSSAVK | 660 |
| KLTFTTVDKT PPVFQTEPTV NSVKETSVGL YANLNEAGTL YWVIVPQGTE YPKPLAGQSG | 720 |
| KVDLTSDTAK LQVAAGMNAL KSGKASMTAG KDVTFTVSGL SKETAYDLYY VAQDKAGNYS | 780 |
| ASVKMITIHT LDPNAPTVTQ EFTRYNADEK DHPLADTDIR LVFSEAVQDA ETNTTLVSLY | 840 |
| EAVTTASTDA EKNDARNRMA NILRNEIVLY ADTGSGLPQE VEVATGFVDK NTADWVIDYR | 900 |
| YAEIKLEQGK TVVTFPTKDV KKESALNLKS GATYYFEVQG IADTSESKNV MGVTKLDPFT | 960 |
| TVFAQVNLSP GSNTETEIDT GSVDDKPELD LYWRMDPVST DKVEDGIRWD MIIWSDTTVE | 1020 |
| FDLYERTGGS GTWTKVNADD KPFTITIDST TKRSGASVQV NDKDNTKPNS PIFQPLKEIK | 1080 |
| EGTVYEYAVH FIKVNGEGDR TTWNGTVQMD VNAIAGSNDN LGRMGVGGWE ESWNNLVGKD | 1140 |
| LTNIGVPSTL TLYKPFRDQT APHFINDRPT FTPGDSVVRM DLMLDRVGTI YWMVAKRGNV | 1200 |
| RTTGMNGEDY SPSGTNPGQY MDLPESGVGK YTLSIATPSY DQVVDPGKYM VSSDVEYGSL | 1260 |
| TCGASVQSVT VENLIPNQDY VAYFVIKGTS QVYSSVYCYR FTTGDVTKPY ITMQELSPNV | 1320 |
| AFTTSEDSDL NYVLFAGTEL PTVFNQKFKQ YVDSSKLAEF STAAGEEADT MTVLDAIMRT | 1380 |
| GTGGYSWFDL YAEPPAKEGE GKKIRDTIEQ IVLRGQGSGG SPGATGNAVT KAGDEIQRDF | 1440 |
| TKDMDPQSAT NYYCIAVAKN VLGGEYAFKA VSNVHIPDKT PPVLYAPGAN GVKKSDGTFS | 1500 |
| GTLTLNFSEN LYWIPENGDT KQICAMLNKN KTATETDAVL EDDGTVTPGK KVLIKHMGGD | 1560 |
| VDTLTPSSTS AISSTFTFSY SGVRIGDTLT LFSDGYIADA RGNSTKTKII LEYQANSTGL | 1620 |
| NGAIAGGWVI ISQ | 1633 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 197 | 309 | 1.7e-19 | 0.9298245614035088 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00395 | SLH | 5.61e-09 | 148 | 189 | 1 | 41 | S-layer homology domain. |
| pfam00395 | SLH | 9.42e-09 | 87 | 128 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 1.93e-07 | 72 | 194 | 567 | 687 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 4.48e-07 | 62 | 198 | 616 | 752 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 3.73e-04 | 41 | 132 | 655 | 748 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA32369.1 | 0.0 | 4 | 1633 | 6 | 1615 |
| QQR07714.1 | 0.0 | 4 | 1633 | 6 | 1615 |
| BCK84769.1 | 0.0 | 8 | 1604 | 17 | 1610 |
| QQR30851.1 | 8.63e-233 | 4 | 1528 | 6 | 1684 |
| ASB41592.1 | 8.63e-233 | 4 | 1528 | 6 | 1684 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 4.08e-08 | 87 | 219 | 26 | 165 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 4.14e-08 | 87 | 219 | 5 | 144 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 4AQ1_A | 7.03e-06 | 96 | 197 | 11 | 109 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 4AQ1_C | 7.03e-06 | 96 | 197 | 11 | 109 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38537 | 6.40e-18 | 34 | 206 | 40 | 219 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P38536 | 4.38e-16 | 35 | 194 | 1688 | 1851 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 1.06e-15 | 35 | 194 | 914 | 1077 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q06852 | 1.32e-13 | 49 | 198 | 2103 | 2264 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
| Q06848 | 6.42e-13 | 49 | 194 | 242 | 394 | Cellulosome-anchoring protein OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=ancA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
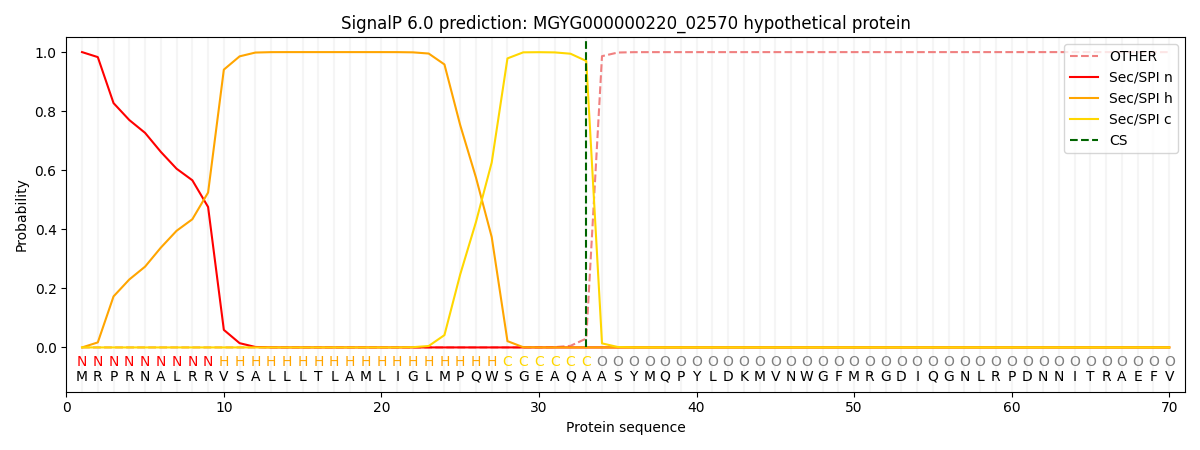
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000314 | 0.998948 | 0.000200 | 0.000198 | 0.000165 | 0.000143 |