You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002863_00106
You are here: Home > Sequence: MGYG000002863_00106
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Agathobaculum desmolans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Butyricicoccaceae; Agathobaculum; Agathobaculum desmolans | |||||||||||
| CAZyme ID | MGYG000002863_00106 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 111; End: 4973 Strand: - | |||||||||||
Full Sequence Download help
| MSHFISHMTR IAKRVCALSL CAALLLGMLP LTTPPAQAGW EDVYLNKLVS WDIMRGDQHG | 60 |
| NLDPGRAITR AEFATMLNRT FGYHKTGTHP FRDVRPADWF YDDISMAYTA GYFQGTSNST | 120 |
| ASPHATLTRE EAATMLCRNL MLQPQSGEDL SFTDSRKASS WSRGYIKAAA KKGVLNGYVD | 180 |
| GTFRPRNRIT RGEVASMLVN VIGTPIQSAG TYSNSVSGNL MVSASGVTLE NMTVTGDLYI | 240 |
| TPGVGLGYVT LNNVTVYGQI IASGAGESNK GDASIILKNC TAPEMIVDSP TNQYVTIRTV | 300 |
| GNTDIDRVYI RTNTYLEGKS TGRHGFAYVE VDGEEGTQLD LAGNMHEVVT MSPKSTINVG | 360 |
| SGTVEKLTVD ENAVGSKVTI AQGAVVEQMN LDTGTTVTGK GDIGTLHVNA PGCTVEMLPD | 420 |
| TIIIRPGITA NINGTIMDST MADQMSDKPR ILSGYPRLSD VAPNQTEASF MTNKPGKLYW | 480 |
| AVRLSGDGAF DANDLIKPPS YGGKIVKSGN LSVKDANDVL SQKITGLQPG ASYVLSAVLV | 540 |
| DARDDRSTVK SVYFTTPDNS KPNFASGYPK TSTIEDTYIA FDVAATKTCT LYWAIYKKGM | 600 |
| PAPTANDFKD GSLSGAISSG KGKLIRSEED NILMGNKDKH AKDALAEVTE YDAYFFLTDT | 660 |
| INDSAVKKVS VKTADRTPPI FRTNYPRISK IDAKALTGES MINEAGKVYW AIVKHGTENY | 720 |
| PQSNPELDEE AQEIDKKLQI KGGMYALKSG SFSAKQDAVS SFNMSGLDAE TAYDIYFVAE | 780 |
| DTAGNLSEIK SILNAKTLDN SRPELVELRF SQANDEGRPL ANTDVTLVFS EDIYNSQAKM | 840 |
| SLVEMYASGK NEFTLAGDKN QTKVQWIDII KGMFEFHNLD NPTDQRKQDL NLGADGRENI | 900 |
| TVKLNEEGQT EVTFANAALK LQSGATYQFV LNNITDSSNN VMPANTKSKE FRVLDAQVDF | 960 |
| NKLQYTAADI DKLNPAGVEA SFSMIPYANS TENVPDGTKY DLLIASDTTI SFKLYRRDLS | 1020 |
| AENPAWEVVT DKKGPVEFTI NKTPAEKWTA ASLNRAKGLE PTDYPQLNQL VKGYEYAVEF | 1080 |
| TSINLDRDRN KWDATVNLYM YCVAGSPNDL QNLTSGGIVE QSFWDSYVIQ QKKLASIGNP | 1140 |
| SDFTIDVRRV NQNAPQFMQG YPRVTPGDSL TRVDYMLDRE ADVYYVVAPV NRLKPETNTP | 1200 |
| NLIIDIGKFQ NDTEAEQALK EPGKREEQGI TAPTTQNIMN PSNYTPANGY QTGKVHYGGT | 1260 |
| GRGSFTVEDL QPNQEYYIYF VLKGSYKDPS PVWCYTFNTS DVVPPVLIAN STGDGQATFS | 1320 |
| VKPAKGQTSV EAEVHWLLYP QATQTGYPPL FTTQINGKTA LEMMESDAFD EWSSDTPTGG | 1380 |
| YDNSTVFKEA LWEIMQGGSN ASPAPDQNGA ERNFSDKDTP KTVIDKNLLK PEPTKYLFIV | 1440 |
| AAKNELGGLP VFRVVKNIQK VNRSGPLVKS MNTTSTYSTK KNWYNGTVTI NFDTELYVYG | 1500 |
| GGSPDMPQPE AYIPTPDRGT TDLFPANSVE YVRVSNLNPT RSIGVQFNNF VVGNSFTITP | 1560 |
| TNGKGNGGYF SNKSGYMRTD GSITFTLQYK RDSDGNPIEE DGANKVEFVL SDGTTSETNP | 1620 |
| 1620 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 197 | 313 | 1.2e-23 | 0.9736842105263158 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00395 | SLH | 4.16e-10 | 152 | 194 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 4.54e-10 | 40 | 202 | 590 | 752 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 1.47e-06 | 63 | 201 | 554 | 690 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 4.27e-06 | 91 | 132 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA32369.1 | 2.67e-311 | 13 | 1497 | 10 | 1496 |
| QQR07714.1 | 2.91e-310 | 13 | 1497 | 10 | 1496 |
| BCK84769.1 | 8.62e-259 | 10 | 1588 | 14 | 1622 |
| AMJ42396.1 | 3.82e-195 | 40 | 1497 | 50 | 1617 |
| QAT42028.1 | 5.29e-192 | 13 | 1497 | 2 | 1398 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GZT_A | 2.98e-09 | 191 | 268 | 33 | 108 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
| 6BT4_A | 3.62e-09 | 84 | 211 | 16 | 143 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 6.74e-09 | 91 | 211 | 5 | 122 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 2.92e-17 | 40 | 202 | 1689 | 1855 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38537 | 8.31e-16 | 38 | 205 | 40 | 213 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P19424 | 2.51e-14 | 37 | 202 | 46 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| Q06852 | 4.90e-11 | 30 | 198 | 2089 | 2260 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
| P06546 | 2.34e-09 | 32 | 259 | 30 | 262 | Middle cell wall protein OS=Brevibacillus brevis (strain 47 / JCM 6285 / NBRC 100599) OX=358681 GN=BBR47_54160 PE=4 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
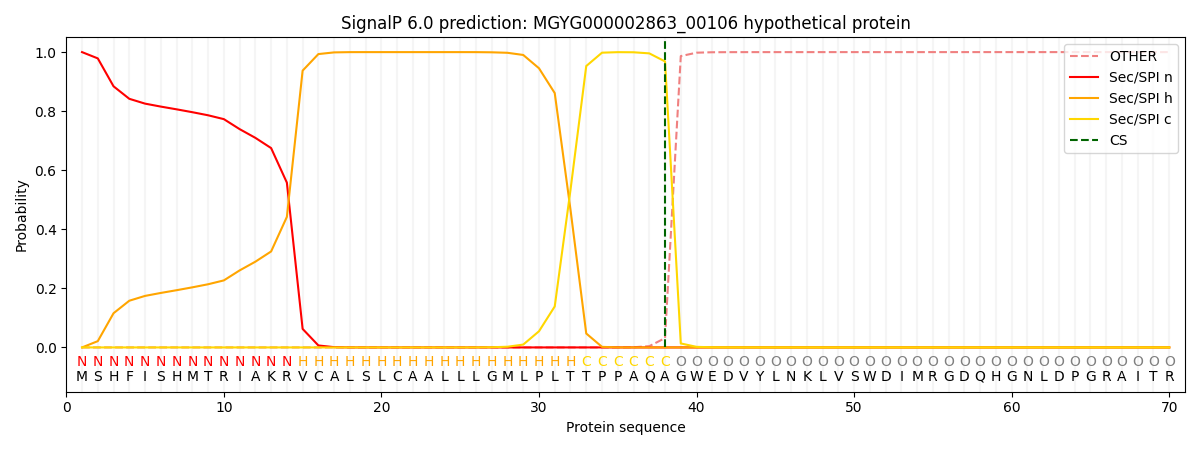
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000392 | 0.998767 | 0.000234 | 0.000212 | 0.000188 | 0.000162 |
