You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004628_00051
You are here: Home > Sequence: MGYG000004628_00051
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
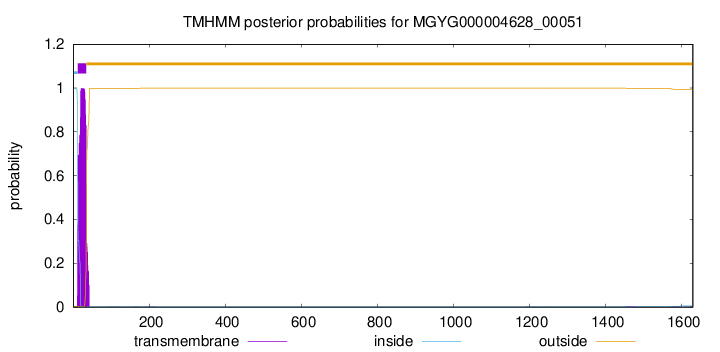
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Lawsonibacter; | |||||||||||
| CAZyme ID | MGYG000004628_00051 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 50771; End: 55663 Strand: + | |||||||||||
Full Sequence Download help
| MTGWMKWMKR LAGRAGTALL VVILAVAMTA AASAASYMQP YLDKVVEWGV MRGDLEGNLN | 60 |
| EDNYITRAEF STMVNRAFGY DKTGPTPFRD VPENSWYAED VGIAYNVGYI AGTSATTFSP | 120 |
| NDNVTREQAA LILARILMLQ PQVGENTMFT DSRSMENWSR GYIASVAQQG LISGYPDGRF | 180 |
| GPKDNLTRGQ AAIILVNALG TPLMEAGDYT LGSVWGNVTI TQSGTTLRNT VIGGDLYITE | 240 |
| GVDLGHVTLE NVTVLGKIVV CGGGTSEGGE DSIILRNVTA PELLVDNLDN HLLSLKVEGD | 300 |
| GKIDQAYIRT SAYLADNTVD GCGIAKITMD GEEDEALTLA GNVKEVINKA PNTSVSLASG | 360 |
| QADTITVDEN AVGSTLDIAA GSRVDNVNLD TGVDVTGGGD IGKLTVNSNG STVSMLPDQI | 420 |
| TIRPGNTANI NGEVMDSAAA AESSADPRLY AGYPKMKDLA PTSGTAVFSA NKKGTVYWAL | 480 |
| TAVTDGSVTA DELVNPSAYN PKVVKSGTVS LTGSGKEGTS NISGLTSDGS YYLSAVFVDA | 540 |
| RDNRSPLKVI SFTTPDNTKP DFASGYPYLS KVTKNSAQVT TMATKSCRLY WAVLPKGASA | 600 |
| PTANDFKANA VSGNLGFGST DVIKNTAYSF DVNNVPLEEL ESYDLYLWLT DVDNGQSSAV | 660 |
| KKLSFTTVDG TPPKFNTEPT INKVDKTSVG LYANLNEAGT LYWVVVEHGT TYPKPLAGQS | 720 |
| GDVDWTSDTA KLQVSAGMNA LKSGKVSMTE GKDVSFTVSG LEAEKAYDLY YVAQDKAGNY | 780 |
| TASIGMIEIH TLDSNAPTVT QEFTKYNGTD TTRPLPDSDV RLVFSESVQT NDTYTPLVDL | 840 |
| YKEVTDAANM GAAEESARQK MAAALSKAIY LYQVSSDGQT EKLEGYTGSD KNTDDWTIDY | 900 |
| RYAVITTEEG KTVVTFPNGK GINLKSGASY YFEIQADSIA DTSSAVNVMG REKLDQFTTV | 960 |
| FAVVNLTARN ENTIEKYVDQ AGTEHELEAK DAIPVDLSWR LSPVSTDKVD DSIDWDMLIW | 1020 |
| SDTSVTLQLF YREVGSSTTT WKLLGEKEIP VPDNTNGYLG VSLTRHFLQN SNNPDFDQLN | 1080 |
| ILKEGTNYEY AISFTKVAGL SDRSTWSQRL NMKVNVVAGS NNDLSFLSGE VTQDNWDEAL | 1140 |
| KTGVTNIGQP SDFSLRKQFT DNVAPTFAGE YPKLDAGDSA VNMSLMLDRP GTVYYVVAPL | 1200 |
| GYVGTTDNKN NIYNNTDAGI TNWENLPENG LDDDINDPDD PYPAILTQPD YLNIVNASSN | 1260 |
| YRNEKIKYGS VTCGSSVETK LVEGLDKQTK YIAYFVLQGT SQTYSRVLAY RFETTDVAKP | 1320 |
| KLTLTDFSPS VNFKTDSTAD VNYALVASNE IPSSLKVDFG SVVDSTRKKE WEDYKKTLDT | 1380 |
| DTNTSVLWAL LTTYKTGYTV FDEFAGDGIR TTVEEFISGV SSMGVDVANR GSTELTQANN | 1440 |
| YSKEQNFTAN MKGATYYYCL ATAVSPLGSE MSFAAVAGIH IRDTEPPKLI SVNTDARPST | 1500 |
| GNLYSGTVTF VFDEPVYQLV NQNGVDQKPM QIWQTKYSIN TDDQKIAVNL PDIISSSLPD | 1560 |
| NFFCETNVRV PSTSLTLTFT NIPLNTTLVL FNNSYICDEN SNSTREILSF TLQSGVPIGN | 1620 |
| IQNGIAFVQQ | 1630 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 202 | 312 | 3.7e-23 | 0.9122807017543859 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 1.22e-13 | 43 | 195 | 541 | 687 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 2.21e-09 | 149 | 191 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 1.55e-08 | 88 | 129 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 4.36e-06 | 62 | 199 | 616 | 752 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 3.93e-05 | 42 | 133 | 655 | 748 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA32369.1 | 0.0 | 36 | 1630 | 36 | 1615 |
| QQR07714.1 | 0.0 | 36 | 1630 | 36 | 1615 |
| BCK84769.1 | 0.0 | 36 | 1517 | 44 | 1536 |
| QQR30851.1 | 5.20e-236 | 36 | 1517 | 38 | 1670 |
| ASB41592.1 | 5.20e-236 | 36 | 1517 | 38 | 1670 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GZT_A | 1.04e-08 | 188 | 264 | 33 | 106 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
| 6BT4_A | 1.22e-08 | 88 | 208 | 26 | 150 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 1.24e-08 | 88 | 208 | 5 | 129 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 4AQ1_A | 4.66e-07 | 97 | 198 | 11 | 109 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 4AQ1_C | 4.66e-07 | 97 | 198 | 11 | 109 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 4.45e-18 | 50 | 199 | 1702 | 1855 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38537 | 2.16e-16 | 36 | 205 | 41 | 216 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P38535 | 9.24e-15 | 50 | 199 | 928 | 1081 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 5.72e-14 | 36 | 199 | 48 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 6.90e-14 | 42 | 195 | 1296 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
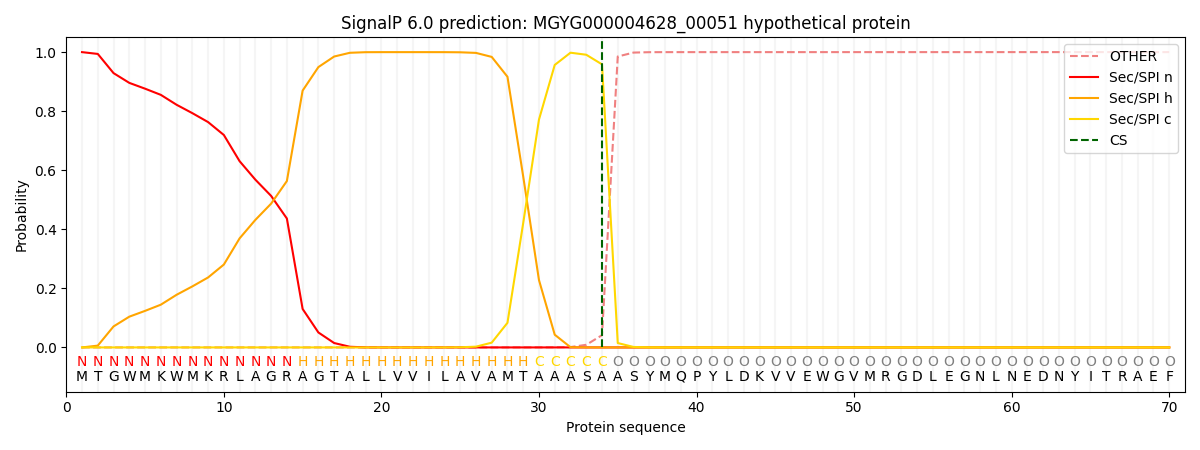
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000739 | 0.998378 | 0.000234 | 0.000218 | 0.000198 | 0.000184 |