You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004493_00403
You are here: Home > Sequence: MGYG000004493_00403
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Spirochaetia; Treponematales; Treponemataceae; Treponema_D; | |||||||||||
| CAZyme ID | MGYG000004493_00403 | |||||||||||
| CAZy Family | CBM61 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53416; End: 53916 Strand: - | |||||||||||
Full Sequence Download help
| MKKTLTAILT LFFAAGLSFG QNLLNDSSFE SGNLSSWNLT GDSADCYAEK NKGNAHSGEW | 60 |
| SYHYWKKTAF TASLDKKITG LSNGTYKLSV WSMGGGGENA IKLFAKGFDN SGKEISAQIK | 120 |
| NTGWKKWKQY SIEIPVQNGE VEIGIFVDAN KENWGNLDDI ELVKLN | 166 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM61 | 21 | 162 | 8.4e-43 | 0.9929078014184397 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02018 | CBM_4_9 | 3.54e-04 | 22 | 131 | 2 | 112 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEB14355.1 | 4.59e-119 | 1 | 166 | 1 | 166 |
| QQA00780.1 | 2.99e-66 | 21 | 164 | 250 | 393 |
| QOS40833.1 | 4.44e-51 | 22 | 165 | 518 | 661 |
| AFK85183.1 | 4.77e-37 | 22 | 165 | 496 | 638 |
| QSI78495.1 | 4.63e-36 | 21 | 164 | 499 | 642 |
SignalP and Lipop Annotations help
This protein is predicted as SP
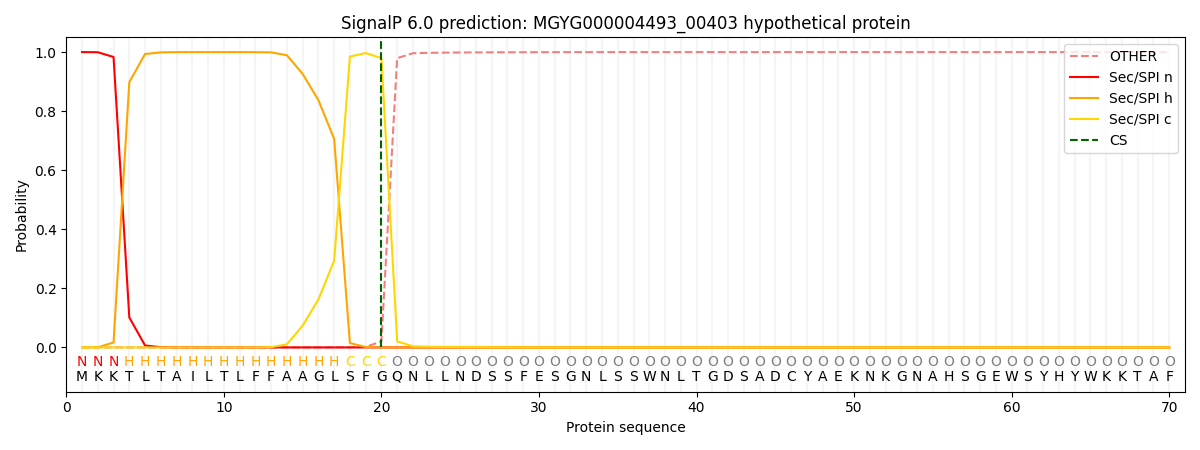
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000395 | 0.998754 | 0.000239 | 0.000213 | 0.000202 | 0.000185 |
