You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000065_00341
You are here: Home > Sequence: MGYG000000065_00341
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerostipes sp900066705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerostipes; Anaerostipes sp900066705 | |||||||||||
| CAZyme ID | MGYG000000065_00341 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | Fructan beta-fructosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 161868; End: 165692 Strand: + | |||||||||||
Full Sequence Download help
| MRKVSPRLLA LMMAGAVTVT SITPVTGYQT ITVNAATDSQ EKEAAQGYQT NLTGFEYKKG | 60 |
| DWKETKDGLY SNAVDKGDCF AFSKTTAKNF VYSTDVTFKR NQGAATLIFR FNNNIDNKEC | 120 |
| YAVNIDGGSH KCKFWRWQEN SDYQLIDEKE VKETDDEKYT LKVVAYDSWI SYYVNDTLIA | 180 |
| STGDYTLQKD DKGQSTVLTE GSLGLLNWNG EMTFQNTYYT ELNDQNTPEL KNISVSSSTG | 240 |
| DVEKAAQFTS TEPIMIQYVK NNAETVDLNI EKKNKNADVQ VEYDGKTYND GKNIPVKVGK | 300 |
| NYITVKSTVQ GENGQTATLT YRVNVHRRAA DKTYYNEAYR NQYHYSVKDG WGNDLNGLVK | 360 |
| YKGTYHMFYQ FYDDTKWGPM HWAHATSKDL IHWEEQPIAL YPDANGAMFS GCIVADEKNT | 420 |
| SGLFGDGNEG GLVALITADG NGQRIKVAYS TDEGKTWKKT NKIAADWSKD PLNNRDFRDP | 480 |
| KVFRWENKWF MVIAGGPLRI YSSDNLTDWK CESTYADLHT ECPDLYPIET KDGVKWVLSR | 540 |
| GGRFYKVGDF KQVDGKWKFV ADEAYKDSDG VMNFGKDSYA AMTYYVQDFG TQENPTIPEI | 600 |
| IEGNWMNTWD DYCNKVADTV GQNFNGTYNL NLKVGLKQEN GKYVLTQTPI SAYESLRDTE | 660 |
| NKISYKDVTI SEDNNLLKDF AKDTYEIVAK FTPSEKTKKV GFRLRKNQND TEYTDVIYDL | 720 |
| QNEKLSIDRS KSGKIISQEF KKINEQTNVK KNEDGSVELH IYVDKASVEI FSNDYTAAGA | 780 |
| NQIFPTPTSL GASVIVEGDP VKADIDIYPM KSIWTDKEEV TDAESVGSMQ NESQSLYVGD | 840 |
| NVELSTYVLP TSVDQTVTWD VTEGKDVVSI KESNGKAVVT ALKSGKATVT AASKLDPSKK | 900 |
| KIFTINVKEN NFKTNIKKFV NISGNWAIDG EVLSDSNQSA NDFYMSEDAI VNEKSTIETD | 960 |
| IAFTNGLVNL IFASSSTDPN GAYCIQFAPD SKNVRLFRMY KDGDIALGEM PSSINDGEYH | 1020 |
| HVKIEKEADA VKVYVDDKEC LNYIFDADKE SDKEFFNIKT GHMGLGLWDG AVSFKNLYVD | 1080 |
| KKEEPTKPTE PTKPTETPKT ISVTLKGNGG KTVTTINKTK AGTKLPGWNK TKYKSAGKKG | 1140 |
| YVFAGWTYKG KVVNKVPASD KNIVLTAKFV KLKVETAKLS SLKGKSGKGT GFVAKSIKYT | 1200 |
| NKYGEKRGFR FRYSTNKKMK AAKYKTTGLA KNTYTKTGLK KGKKYYVQVR YYYYDSTNKK | 1260 |
| VYGTYSKVKS VKAY | 1274 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH32 | 344 | 648 | 1.7e-64 | 0.9931740614334471 |
| CBM66 | 50 | 216 | 4.3e-45 | 0.9870967741935484 |
| CBM66 | 914 | 1076 | 5e-35 | 0.9870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00640 | Glyco_32 | 3.40e-101 | 344 | 774 | 1 | 436 | Glycosyl hydrolases family 32. |
| cd18622 | GH32_Inu-like | 1.76e-95 | 349 | 636 | 1 | 289 | glycoside hydrolase family 32 protein such as Aspergillus ficuum endo-inulinase (Inu2). This subfamily of glycosyl hydrolase family GH32 includes endo-inulinase (inu2, EC 3.2.1.7), exo-inulinase (Inu1, EC 3.2.1.80), invertase (EC 3.2.1.26), and levan fructotransferase (LftA, EC 4.2.2.16), among others. These enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| COG1621 | SacC | 1.69e-86 | 329 | 812 | 18 | 486 | Sucrose-6-phosphate hydrolase SacC, GH32 family [Carbohydrate transport and metabolism]. |
| cd08996 | GH32_FFase | 2.40e-63 | 350 | 630 | 1 | 275 | Glycosyl hydrolase family 32, beta-fructosidases. Glycosyl hydrolase family GH32 cleaves sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| pfam00251 | Glyco_hydro_32N | 1.60e-61 | 344 | 649 | 1 | 308 | Glycosyl hydrolases family 32 N-terminal domain. This domain corresponds to the N-terminal domain of glycosyl hydrolase family 32 which forms a five bladed beta propeller structure. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQP39972.1 | 0.0 | 1 | 1274 | 1 | 1303 |
| CBL39539.1 | 0.0 | 1 | 1273 | 1 | 1296 |
| ACV51683.1 | 0.0 | 60 | 1079 | 168 | 1175 |
| SQG29543.1 | 0.0 | 36 | 1085 | 141 | 1177 |
| QLB51563.1 | 0.0 | 36 | 1085 | 144 | 1179 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1Y4W_A | 6.19e-53 | 335 | 814 | 3 | 517 | Crystalstructure of exo-inulinase from Aspergillus awamori in spacegroup P21 [Aspergillus awamori],1Y9G_A Crystal structure of exo-inulinase from Aspergillus awamori complexed with fructose [Aspergillus awamori],1Y9M_A Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121 [Aspergillus awamori] |
| 7VCO_A | 1.16e-36 | 343 | 813 | 29 | 488 | ChainA, Sucrose-6-phosphate hydrolase [Frischella perrara],7VCP_A Chain A, Sucrose-6-phosphate hydrolase [Frischella perrara] |
| 4EQV_A | 7.50e-35 | 344 | 610 | 12 | 292 | Structureof Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_B Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_C Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_D Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_E Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_F Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_G Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_H Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C] |
| 3KF3_A | 1.07e-32 | 344 | 590 | 14 | 278 | ChainA, Invertase [Schwanniomyces occidentalis],3KF3_B Chain B, Invertase [Schwanniomyces occidentalis] |
| 3KF5_A | 1.11e-32 | 344 | 590 | 17 | 281 | ChainA, Invertase [Schwanniomyces occidentalis],3KF5_B Chain B, Invertase [Schwanniomyces occidentalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q03174 | 0.0 | 50 | 1088 | 152 | 1180 | Fructan beta-fructosidase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=fruA PE=2 SV=2 |
| P05656 | 1.26e-76 | 329 | 1077 | 24 | 673 | Levanase OS=Bacillus subtilis (strain 168) OX=224308 GN=sacC PE=1 SV=1 |
| O31411 | 7.41e-60 | 38 | 818 | 45 | 883 | Levanase (Fragment) OS=Bacillus sp. (strain L7) OX=62626 PE=1 SV=2 |
| E1ABX2 | 4.05e-54 | 335 | 814 | 22 | 536 | Extracellular exo-inulinase inuE OS=Aspergillus ficuum OX=5058 GN=exoI PE=1 SV=1 |
| Q76HP6 | 4.05e-54 | 335 | 814 | 22 | 536 | Extracellular exo-inulinase inuE OS=Aspergillus niger OX=5061 GN=inuE PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
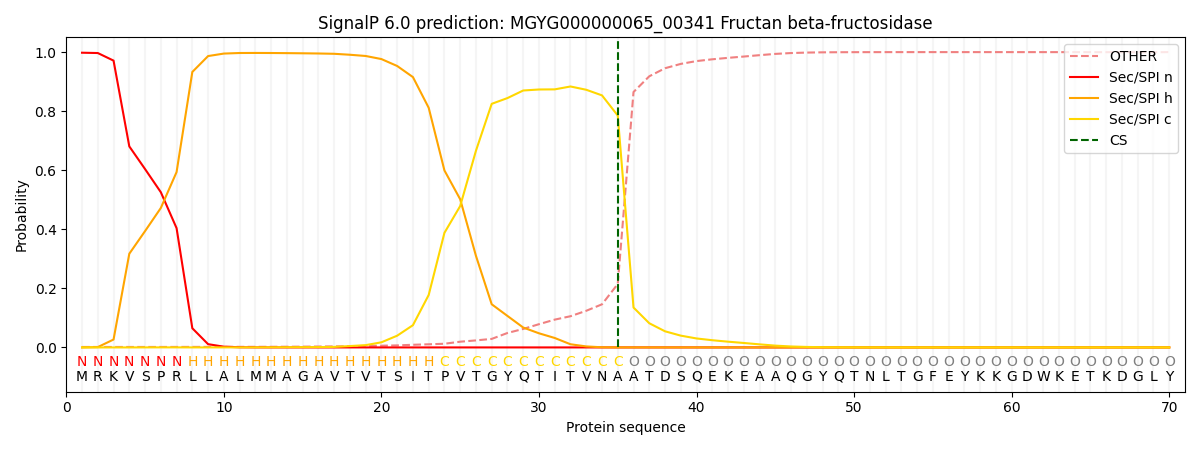
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002732 | 0.995413 | 0.000709 | 0.000633 | 0.000270 | 0.000218 |
