You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000069_00543
You are here: Home > Sequence: MGYG000000069_00543
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
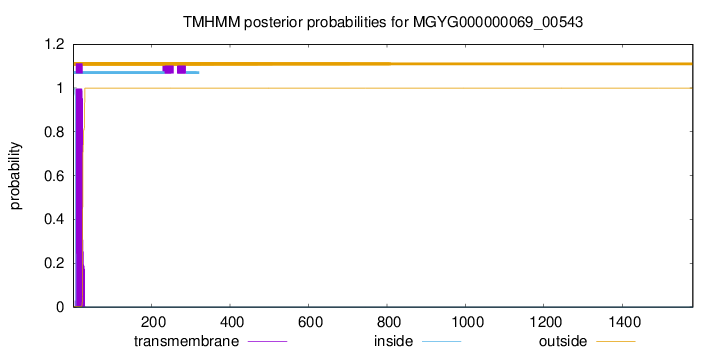
TMHMM annotations
Basic Information help
| Species | Clostridium_A leptum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Clostridium_A; Clostridium_A leptum | |||||||||||
| CAZyme ID | MGYG000000069_00543 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 572182; End: 576924 Strand: - | |||||||||||
Full Sequence Download help
| MRKIGKIIFS FLFVCCTVIG FYITDQSFGM EASAAGNPPQ APERLKVNLL DKPLGVSKDG | 60 |
| LRFSWSFEDP DANEYQSAYR IVVASSKAQF LQKEYVYDSG WSDASASSGV EPAGLPECLT | 120 |
| DNGLYYWAVQ VRDSQGLESE LSQPEMLVTS VGDQWTNKNG IWGSSSQKFV FLRNKLSLDK | 180 |
| PVEKVIASVT AASTETTQQY VYQFYVNGQL VGLGPSVKNL SDLYYNTYDI TSLLNQGENI | 240 |
| LGAVCYAEDK QGFLCQITAF YEDGTKEVLC NSGSNPSSWQ ALDANEIYGY QGKSIASYYH | 300 |
| ASPENLNGTK FPYGWNQAEF QGTGWKSALS SGSIEEKNQG ELTPYPSGNM TRYQQPAASV | 360 |
| TRLSDGSYVV DLGKEIIGSI RLTVDSPKQQ TVTLLYGEER NNDGTVKSKM RTGNVYEEQW | 420 |
| VLKKGEQTLE GIGMKCFRYI QIKNFPAALT AEQITGLTIR QEFDDSQSSF SSSNQMLNEI | 480 |
| YEMTKYTVKT AAQSQYVDSQ SRERKTYEGD ILVQMMSSFS FLDDYSLARH SLDYVVNNPT | 540 |
| WPAEYQLFAI TAAWQEYLYS GDIAFLRDHY DQLKNALYDS SYNSNLGLVH NPGKVVMVDW | 600 |
| PYSQRDSYEV SNTAYNTAFN AVCSAAYRDM GNIAAALGYG DDASFLKERS DVIKQNMISR | 660 |
| LYDQNTGRFY DGLTEAGAVV NHCAQHATAF SLACGIYADQ AMADRMSATI VADGTIRMSV | 720 |
| YGSYFLLDGL YQSGSGTLAR QFMSNPDTQY SSNSWAYMLK KLGATMSTEA WSPEAKGNMT | 780 |
| FSHAWGSSPA SQIVRGMFGI KPTAPGFSQF EVKVQPGGLT EGAVEIPTVK GTIPVSFRLA | 840 |
| QDGVITARVS VPANTQAQVL LPANADGSRS VTVNGTDTQA EVQQNFVKVS LGSGTYELVY | 900 |
| DTGTAPDPSE ITIPPVVNAE AYVGGLYFWQ EPVTMDGVTC GTEGRGLPLN GLRFTLSGNG | 960 |
| ISGGISSSVN LIKNGWQGWK EAGELNGNAG NGQAVQAVRL KLTGEAAQQY DLYYRVHSKT | 1020 |
| YGWLDWAKNG QIAGTIGLDK QMEAIQIRIV KKDGGAPGST TRPFVQKSQD VLTYQTHVQS | 1080 |
| YGWQNDVGQG KISGTVGQAK RLEAIRINVH STEGSIRYRT HVQTYGWQDW AYDGEESGTV | 1140 |
| GQAKRLEAIQ IELTGGLAEK YDVYYRVHSQ TYGWLGWVKN GEPAGTEGLA KRLEGIQITL | 1200 |
| VEKGGSAPGM YGGGFIKPGL GTKDGVNYKT HIESYGWQSW RSNGESSGTM GEAKRLEGIC | 1260 |
| IQVQNPGAAG DIEYQTHVQT YGWQNWVKNG AVSGTQGQAK RLEAIKIRLT GALAEKYDVY | 1320 |
| YRVHAQTYGW LDWAKNGEAA GTEGLAKRLE AIEIRLVEKG GSAPGSTERP YVTPPPATVD | 1380 |
| GINYQTHVQI YGWQAWKANG AVSGTSGEAK RLESIRILLQ NPGYSGSVEY QTHVQTYGWQ | 1440 |
| NWVKDGAESG TSGQAKRLEG IKIRLTGELA EHYDIYYRVH VQTYGWQDWV KNGEMAGTSG | 1500 |
| EAKRLEAIEI KLVEKPKPVP PEVSESQAPS ENVSTDPALE SSSETESLAS QSSNSVEAEI | 1560 |
| ENGSLSQSQS SVESSKFTDD | 1580 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 360 | 860 | 2.1e-104 | 0.996031746031746 |
| CBM67 | 161 | 333 | 2.3e-21 | 0.9034090909090909 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17389 | Bac_rhamnosid6H | 2.60e-25 | 474 | 790 | 10 | 333 | Bacterial alpha-L-rhamnosidase 6 hairpin glycosidase domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| COG5492 | YjdB | 2.44e-23 | 1219 | 1465 | 19 | 306 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 2.66e-21 | 1378 | 1516 | 22 | 161 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 1.61e-19 | 1070 | 1309 | 24 | 306 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam07538 | ChW | 2.00e-18 | 1277 | 1311 | 1 | 35 | Clostridial hydrophobic W. A novel extracellular macromolecular system has been proposed based on the proteins containing ChW repeats. ChW stands for Clostridial hydrophobic with conserved W (tryptophan). This repeat was originally described in Clostridium acetobutylicum but is also found in other Gram-positive bacteria including Enterococcus faecalis, Streptococcus agalactiae and Streptomyces coelicolor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQW20897.1 | 2.87e-203 | 46 | 898 | 35 | 871 |
| AFS01190.1 | 2.39e-182 | 42 | 902 | 11 | 907 |
| QUX04741.1 | 3.49e-181 | 42 | 902 | 11 | 907 |
| APR07480.1 | 4.87e-181 | 42 | 900 | 11 | 905 |
| AEB74357.1 | 7.08e-180 | 42 | 902 | 11 | 907 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6I60_A | 5.60e-33 | 32 | 896 | 22 | 937 | Structureof alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12],6I60_B Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12] |
| 6GSZ_A | 2.84e-23 | 54 | 862 | 16 | 847 | Crystalstructure of native alfa-L-rhamnosidase from Aspergillus terreus [Aspergillus terreus] |
| 3W5M_A | 4.48e-23 | 189 | 905 | 316 | 1035 | CrystalStructure of Streptomyces avermitilis alpha-L-rhamnosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3W5N_A Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 2OKX_A | 1.72e-16 | 365 | 867 | 436 | 947 | Crystalstructure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A [Bacillus sp. GL1],2OKX_B Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A [Bacillus sp. GL1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM26 | 1.66e-54 | 365 | 861 | 604 | 1136 | Bifunctional sulfatase/alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22250 PE=1 SV=2 |
| T2KPL4 | 2.25e-29 | 43 | 901 | 33 | 950 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22170 PE=2 SV=1 |
| Q82PP4 | 3.19e-22 | 189 | 896 | 316 | 1026 | Alpha-L-rhamnosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=SAVERM_828 PE=1 SV=1 |
| T2KNB2 | 1.35e-18 | 53 | 897 | 47 | 919 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22090 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
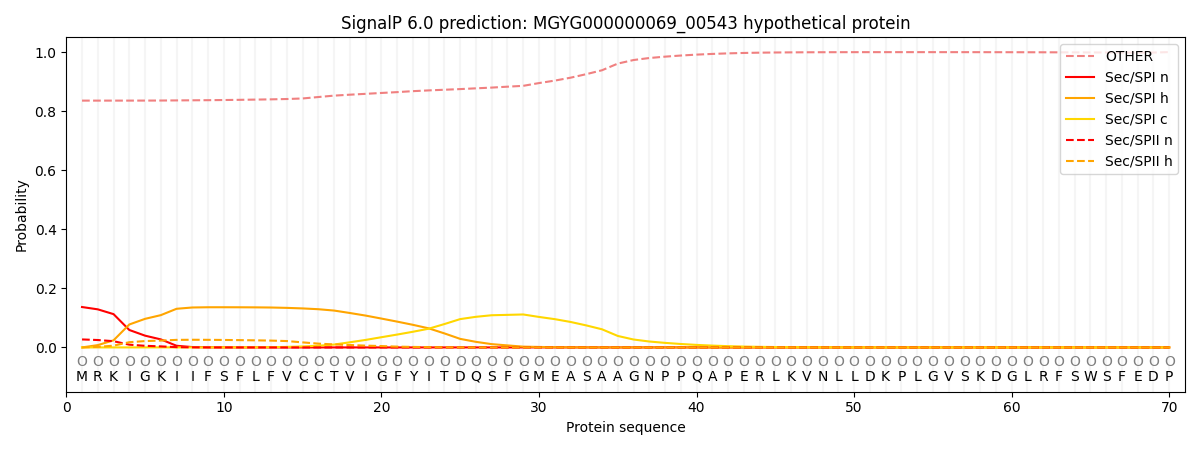
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.841786 | 0.128871 | 0.027856 | 0.000297 | 0.000222 | 0.000975 |