You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004207_00108
You are here: Home > Sequence: MGYG000004207_00108
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
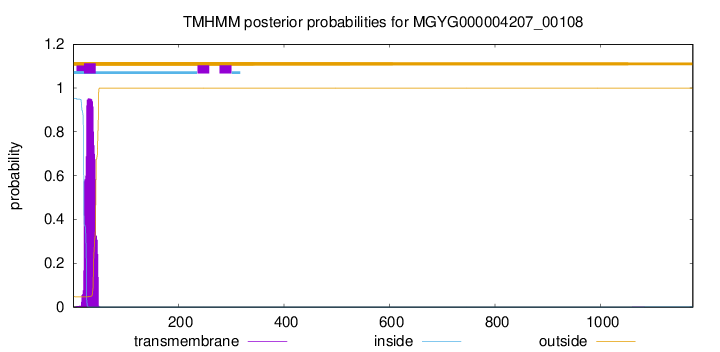
TMHMM annotations
Basic Information help
| Species | UBA6398 sp003150315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA6398; UBA6398 sp003150315 | |||||||||||
| CAZyme ID | MGYG000004207_00108 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 122788; End: 126315 Strand: + | |||||||||||
Full Sequence Download help
| MRRATMFILY VLEMRMTIYN VFPKKYIAII IMLLAAVMPS AATDIITCSE EVENGMWVCF | 60 |
| RKTFELAEAP SENTLRISAD SKYRLYVNGY IVVREGQLKR GPNPHDSYID SLGIKNLRKG | 120 |
| KNTVAVLVWY FGKGGYSHRS SSHAGLYFDL SGKAFRVSSD SSWKSRLHPA YYIPTGQKPN | 180 |
| ARLAESNIGY DARQELQSFA DTLYNDSAWA NAVQMNADSA GWGAFRSRPI PFFWHSEVKD | 240 |
| YVSFHQTGNK IYCYLPYNAQ VTPVLHIKAN AAGKTIGIQS DCYPKSGTDY VMRYEYKTKT | 300 |
| GEQEYEFPAW INGHVIIYTV PEGVEVIRLA YRETGYDCSL DGSFDSSDET LNKLWRKAQR | 360 |
| TLYVTMRDNF MDCPDRERAQ YIGDVTNELA EVPYAMSLSA SQLIRKCARE FADWQKKDSV | 420 |
| LYAPVPSGEW NNELPQQSLS FCGLGLWDYY RYYGDANTIE YVYPAVKKYL HLWKVQPDSL | 480 |
| VNYRSGSWDW GDWGTNVDLK TLNQAWYSVT LGYFSKMARL NGDDNESAWA DTVRRNLNSA | 540 |
| IFSKYWRGKA FNSSKVSKND DRAQALAILA GAAPPHVWDD VRKVFAQTEY ASPYMERFVL | 600 |
| QALCEMNNTQ DALNRMRRRY RVMADSVYTT LWERFEPAPN STYNHAWSGA PLIILSRYVC | 660 |
| GITPLQPLFK EFQVKPQLCD LDYIRTAVPT PGGRISLYVS KKDGYRMQLD VPKGSNARVL | 720 |
| LPSDYLGYHL NGKEIKLKLS EDTAFYELNL SHGYYVVEAL INEADTVGAG ARLANAMARL | 780 |
| RYLRGNVSAG NGVGNAPAAY VSRFAAEADS LMASVEPGND VYETVFRLRE AENLCSALCD | 840 |
| SLFLPSPGKW YEISAASTGA LLETDNVQRS RFPLGGVWIA GNAEEGHLFG MYNAASGDVF | 900 |
| SEFPMGVFPA KDGKVALTDD SGGKYLRVLP EGFSEWTETG IDSLARHEFS FSFDAVEKPT | 960 |
| LCFSDTMDGG AWKAVILPTR IDSVDGAGVS LYTVCGLMKD ASGKLAGIKV SRLMRDSIFL | 1020 |
| SAGQPVIYHV DSAAGKTTEV SFYAQPHTPL SFPGKRSNGL VGVSDTTVVK QSGMLAFVNG | 1080 |
| VLCKADAPFT VMPRDCYINP KYVSELPTGA DEIIFLGGSD TPDAVSLVRL SHKKEYVDVY | 1140 |
| TPDGLRVKRN VKRSKLSSTL RKGIYIIDGK KRVIK | 1175 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 295 | 720 | 1.8e-83 | 0.878968253968254 |
| CBM67 | 50 | 216 | 9.1e-32 | 0.9090909090909091 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17389 | Bac_rhamnosid6H | 9.22e-35 | 342 | 659 | 4 | 340 | Bacterial alpha-L-rhamnosidase 6 hairpin glycosidase domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam17390 | Bac_rhamnosid_C | 1.02e-09 | 661 | 724 | 1 | 66 | Bacterial alpha-L-rhamnosidase C-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. L-Rhamnose is abundant in biomass as a common constituent of glycolipids and glycosides, such as plant pigments, pectic polysaccharides, gums or biosurfactants. Some rhamnosides are important bioactive compounds. For example, terpenyl glycosides, the glycosidic precursor of aromatic terpenoids, act as important flavouring substances in grapes. Other rhamnosides act as cytotoxic rhamnosylated terpenoids, as signal substances in plants or play a role in the antigenicity of pathogenic bacteria. |
| pfam08531 | Bac_rhamnosid_N | 1.45e-09 | 69 | 230 | 2 | 168 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH54696.1 | 1.13e-218 | 24 | 734 | 5 | 719 |
| QNL36670.1 | 3.01e-212 | 44 | 734 | 23 | 719 |
| ACU03186.1 | 7.23e-181 | 42 | 697 | 34 | 701 |
| BCI61942.1 | 2.12e-179 | 40 | 756 | 23 | 755 |
| QLG44645.1 | 2.00e-176 | 56 | 741 | 40 | 737 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CIH_A | 3.57e-21 | 342 | 702 | 302 | 679 | Crystalstructure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPL4 | 2.01e-07 | 60 | 733 | 187 | 919 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22170 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
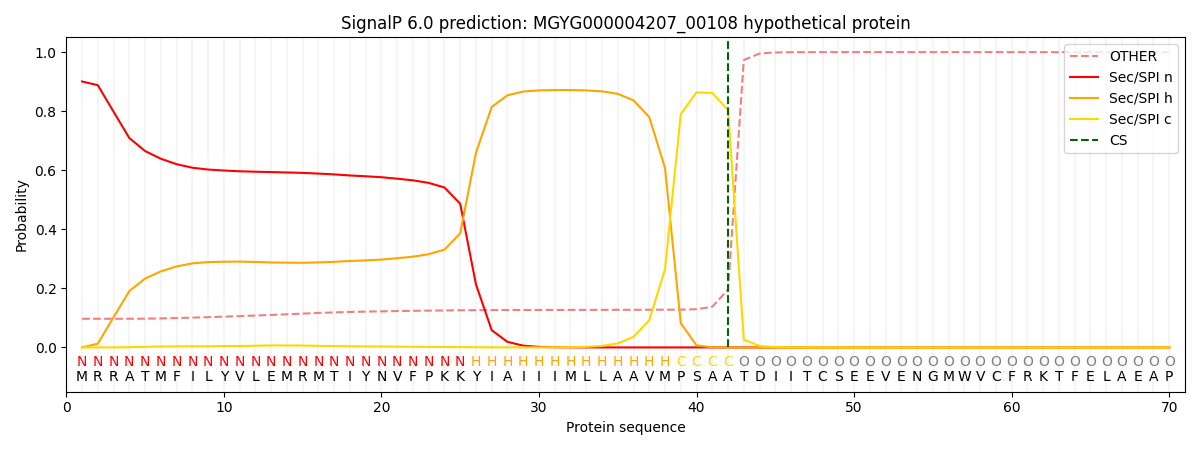
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.102632 | 0.893238 | 0.003220 | 0.000318 | 0.000284 | 0.000308 |