You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000358_00505
You are here: Home > Sequence: MGYG000000358_00505
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1436 sp002329395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Elusimicrobiota; Elusimicrobia; Elusimicrobiales; Elusimicrobiaceae; UBA1436; UBA1436 sp002329395 | |||||||||||
| CAZyme ID | MGYG000000358_00505 | |||||||||||
| CAZy Family | CE11 | |||||||||||
| CAZyme Description | UDP-3-O-acyl-N-acetylglucosamine deacetylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6169; End: 6993 Strand: - | |||||||||||
Full Sequence Download help
| MRKTLASPAV VSGIGLHTGV PATMTFKPAD NGITFLRTDI KDAKPIQARL DNVGSTLRGT | 60 |
| NLKNETAEVW TVEHALSALH ALGITDAAVE MDGPEPPVTD GASDQYVDAL QKAGLTELDA | 120 |
| EQPLLNIKQK ITYSCGSISY SAEPADKLTF TFLFLHKHPL VSRQEFTLEF TPENYIKEIA | 180 |
| RARTFGFEEE LAFLKAHGLA KGGNLENAVI VGKDKFLNEL RYRDELVRHK ILDLVGDISL | 240 |
| TGRKLPPLKI TCACGGHKHN VIFAKILLAN SEDK | 274 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE11 | 3 | 268 | 1.3e-85 | 0.988929889298893 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03331 | LpxC | 1.03e-121 | 2 | 269 | 1 | 271 | UDP-3-O-acyl N-acetylglycosamine deacetylase. The enzymes in this family catalyze the second step in the biosynthetic pathway for lipid A. |
| PRK13186 | lpxC | 1.17e-112 | 1 | 272 | 3 | 277 | UDP-3-O-acyl-N-acetylglucosamine deacetylase. |
| COG0774 | LpxC | 8.05e-95 | 1 | 272 | 3 | 280 | UDP-3-O-acyl-N-acetylglucosamine deacetylase [Cell wall/membrane/envelope biogenesis]. |
| PRK13188 | PRK13188 | 6.17e-85 | 1 | 267 | 4 | 298 | bifunctional UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase/(3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase; Reviewed |
| PRK13187 | PRK13187 | 8.19e-26 | 4 | 269 | 13 | 292 | UDP-3-O-acyl N-acetylglycosamine deacetylase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACC97643.1 | 1.02e-106 | 2 | 268 | 3 | 270 |
| AVQ26965.1 | 2.24e-64 | 2 | 267 | 3 | 271 |
| SQJ09694.1 | 2.24e-64 | 2 | 267 | 3 | 271 |
| AYF45914.1 | 3.98e-64 | 2 | 267 | 4 | 273 |
| BBA50466.1 | 5.07e-63 | 2 | 267 | 3 | 271 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5U39_A | 1.24e-55 | 2 | 245 | 6 | 255 | Pseudomonasaeruginosa LpxC in complex with CHIR-090 [Pseudomonas aeruginosa PAO1] |
| 4J3D_A | 1.31e-55 | 2 | 245 | 4 | 253 | Pseudomonasaeruginosa LpxC in complex with a hydroxamate inhibitor [Pseudomonas aeruginosa PAO1],4J3D_B Pseudomonas aeruginosa LpxC in complex with a hydroxamate inhibitor [Pseudomonas aeruginosa PAO1] |
| 5U3B_A | 1.38e-55 | 2 | 245 | 4 | 253 | Pseudomonasaeruginosa LpxC in complex with NVS-LPXC-01 [Pseudomonas aeruginosa PAO1],5U3B_B Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 [Pseudomonas aeruginosa PAO1],6MAE_A CHAIN A. UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase PA-LPXC Complexed with (R)-3-((S)-3-(4-(cyclopropylethynyl)phenyl)-2-oxooxazolidin-5-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)propenamide [Pseudomonas aeruginosa PAO1] |
| 7K99_A | 1.54e-55 | 2 | 245 | 4 | 253 | CrystalStructure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 [Pseudomonas aeruginosa PAO1],7K99_C Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 [Pseudomonas aeruginosa PAO1],7K9A_A Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor [Pseudomonas aeruginosa PAO1],7K9A_C Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor [Pseudomonas aeruginosa PAO1] |
| 6MO4_A | 1.54e-55 | 2 | 245 | 8 | 257 | Co-Crystalstructure of P. aeruginosa LpxC-50067 complex [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B5EFL8 | 8.08e-59 | 2 | 245 | 4 | 251 | UDP-3-O-acyl-N-acetylglucosamine deacetylase OS=Citrifermentans bemidjiense (strain ATCC BAA-1014 / DSM 16622 / JCM 12645 / Bem) OX=404380 GN=lpxC PE=3 SV=1 |
| C6E4T9 | 3.21e-58 | 2 | 245 | 4 | 251 | UDP-3-O-acyl-N-acetylglucosamine deacetylase OS=Geobacter sp. (strain M21) OX=443144 GN=lpxC PE=3 SV=1 |
| B9L6E8 | 1.98e-56 | 2 | 267 | 3 | 269 | UDP-3-O-acyl-N-acetylglucosamine deacetylase OS=Nautilia profundicola (strain ATCC BAA-1463 / DSM 18972 / AmH) OX=598659 GN=lpxC PE=3 SV=1 |
| Q4K6J9 | 2.69e-56 | 2 | 245 | 4 | 253 | UDP-3-O-acyl-N-acetylglucosamine deacetylase OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=lpxC PE=3 SV=1 |
| B3PCL2 | 3.80e-56 | 2 | 245 | 4 | 253 | UDP-3-O-acyl-N-acetylglucosamine deacetylase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=lpxC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
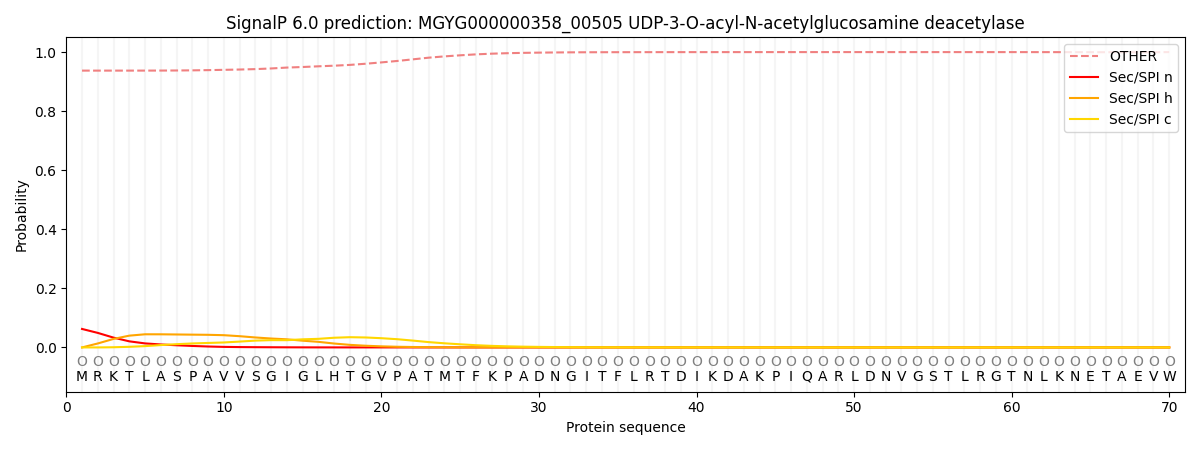
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.942297 | 0.057210 | 0.000254 | 0.000102 | 0.000056 | 0.000104 |
