You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002595_00716
You are here: Home > Sequence: MGYG000002595_00716
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
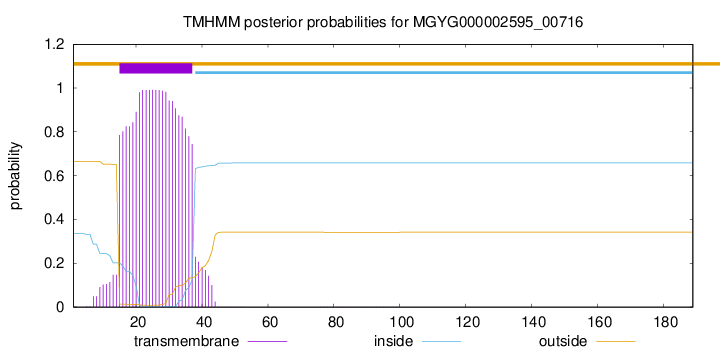
TMHMM annotations
Basic Information help
| Species | CAG-83 sp902388735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-83; CAG-83 sp902388735 | |||||||||||
| CAZyme ID | MGYG000002595_00716 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | Peptidoglycan-N-acetylmuramic acid deacetylase PdaC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3759; End: 4328 Strand: - | |||||||||||
Full Sequence Download help
| MSRKPVRVFL IPKSIALAAC ALLAAAAIFG AVHVPAAITA VTAARQLPIY SVDRPDEGGK | 60 |
| KYCAISFDAA WGNEDTQMLI GILNQYQVKA TFFLVGDWVD KYPESVQALH EAGMEVMNHS | 120 |
| NHHDHYNSLS ADQIVADVTA CNDKVKAITG IAPTLIRCPY GEYDDHVISA IRSLGMEPIQ | 180 |
| WDVEPPAAE | 189 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 57 | 177 | 1.1e-30 | 0.9076923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10917 | CE4_NodB_like_6s_7s | 7.67e-45 | 61 | 185 | 1 | 125 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| cd10950 | CE4_BsYlxY_like | 2.94e-39 | 60 | 183 | 5 | 128 | Putative catalytic NodB homology domain of uncharacterized protein YlxY from Bacillus subtilis and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by Bacillus subtilis putative polysaccharide deacetylase BsYlxY, encoded by the ylxY gene, which is a member of the carbohydrate esterase 4 (CE4) superfamily. Although its biological function still remains unknown, BsYlxY shows high sequence homology to the catalytic domain of Bacillus subtilis pdaB gene encoding a putative polysaccharide deacetylase (BsPdaB), which is essential for the maintenance of spores after the late stage of sporulation and is highly conserved in spore-forming bacteria. However, disruption of the ylxY gene in B. subtilis did not cause any sporulation defect. Moreover, the Asp residue in the classical His-His-Asp zinc-binding motif of CE4 esterases is mutated to a Val residue in this family. Other catalytically relevant residues of CE4 esterases are also not conserved, which suggest that members of this family may be inactive. |
| TIGR02764 | spore_ybaN_pdaB | 6.06e-39 | 60 | 185 | 5 | 130 | polysaccharide deacetylase family sporulation protein PdaB. This model describes the YbaN protein family, also called PdaB and SpoVIE, of Gram-positive bacteria. Although ybaN null mutants have only a mild sporulation defect, ybaN/ytrI double mutants show drastically reducted sporulation efficiencies. This synthetic defect suggests the role of this sigmaE-controlled gene in sporulation had been masked by functional redundancy. Members of this family are homologous to a characterized polysaccharide deacetylase; the exact function this protein family is unknown. [Cellular processes, Sporulation and germination] |
| pfam01522 | Polysacc_deac_1 | 4.02e-37 | 57 | 179 | 3 | 124 | Polysaccharide deacetylase. This domain is found in polysaccharide deacetylase. This family of polysaccharide deacetylases includes NodB (nodulation protein B from Rhizobium) which is a chitooligosaccharide deacetylase. It also includes chitin deacetylase from yeast, and endoxylanases which hydrolyzes glucosidic bonds in xylan. |
| COG0726 | CDA1 | 3.66e-33 | 55 | 185 | 59 | 190 | Peptidoglycan/xylan/chitin deacetylase, PgdA/CDA1 family [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCK79020.1 | 3.98e-103 | 1 | 184 | 1 | 184 |
| BCK80391.1 | 1.29e-80 | 23 | 184 | 20 | 177 |
| QUO36307.1 | 1.04e-76 | 7 | 184 | 2 | 175 |
| QNL43472.1 | 1.44e-75 | 22 | 184 | 17 | 175 |
| ANU39676.1 | 1.50e-70 | 13 | 184 | 9 | 175 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6HM9_A | 9.82e-17 | 60 | 184 | 84 | 208 | Crystalstructure of a BA3943 mutant,a CE4 family pseudoenzyme with restored enzymatic activity. [Bacillus anthracis] |
| 7FBW_A | 1.30e-16 | 61 | 185 | 117 | 241 | ChainA, Predicted xylanase/chitin deacetylase [Caldanaerobacter subterraneus subsp. tengcongensis MB4] |
| 6HPA_A | 3.41e-16 | 60 | 184 | 85 | 209 | Crystalstructure of a BA3943 mutant,a CE4 family pseudoenzyme [Bacillus anthracis] |
| 6H8L_A | 3.77e-16 | 60 | 185 | 9 | 131 | Structureof peptidoglycan deacetylase PdaC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],6H8L_B Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 7BKF_A | 1.91e-15 | 60 | 184 | 85 | 209 | ChainA, Putative polysaccharide deacetylase [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P50850 | 5.59e-15 | 48 | 184 | 121 | 253 | Uncharacterized protein YlxY OS=Bacillus subtilis (strain 168) OX=224308 GN=ylxY PE=3 SV=2 |
| O34798 | 1.34e-14 | 60 | 185 | 277 | 399 | Peptidoglycan-N-acetylmuramic acid deacetylase PdaC OS=Bacillus subtilis (strain 168) OX=224308 GN=pdaC PE=1 SV=1 |
| P54865 | 5.08e-14 | 62 | 184 | 357 | 479 | Bifunctional xylanase/deacetylase OS=Cellulomonas fimi OX=1708 GN=xynD PE=1 SV=1 |
| Q04729 | 4.21e-13 | 60 | 183 | 66 | 190 | Uncharacterized 30.6 kDa protein in fumA 3'region OS=Geobacillus stearothermophilus OX=1422 PE=3 SV=1 |
| P50354 | 5.79e-12 | 49 | 185 | 7 | 144 | Chitooligosaccharide deacetylase OS=Rhizobium galegae OX=399 GN=nodB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
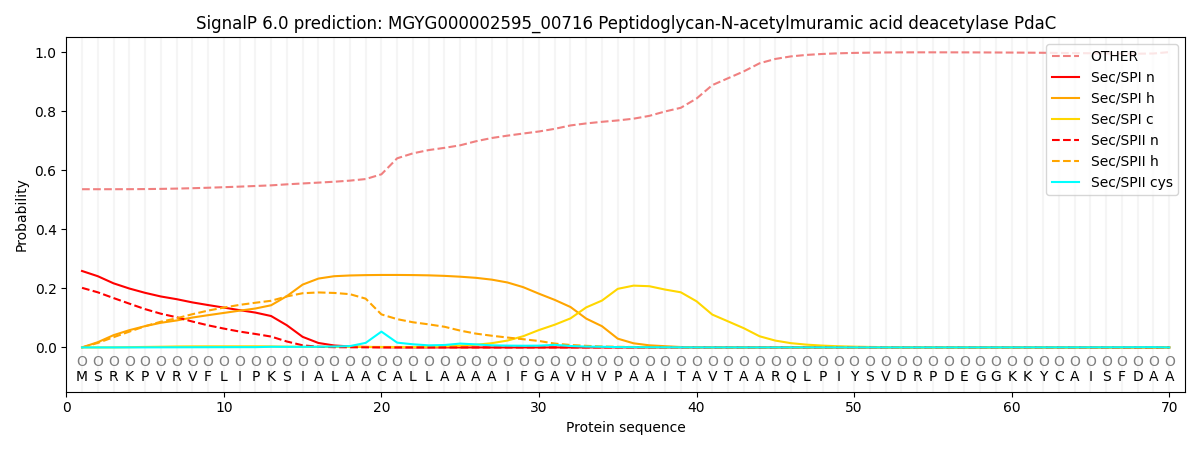
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.546830 | 0.241721 | 0.204222 | 0.001184 | 0.000825 | 0.005221 |