You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004594_00068
You are here: Home > Sequence: MGYG000004594_00068
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
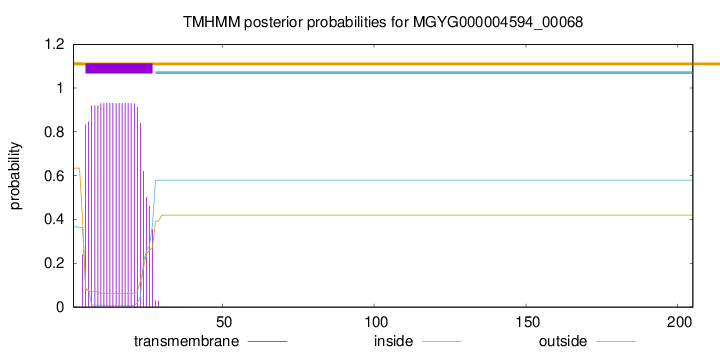
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; TANB77; CAG-508; UMGS1663; | |||||||||||
| CAZyme ID | MGYG000004594_00068 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 73255; End: 73872 Strand: + | |||||||||||
Full Sequence Download help
| MKKYYYILEL IVTLVFTISV ISLNSNYDAV QTSGTIATLS TKKIEWGIKR NNNHEQPDLG | 60 |
| TVNKKIIDEA KGIAMGNKES KKVYLTFDEG YEAGYTEKIL EVLKQNDVKA CFFVTAHYVN | 120 |
| TSSDLVQKMI DDGHTIGNHT VNHKSMPSLT IEQIKDEVMK LHTGVYEKFG YEMKYIRPPK | 180 |
| GEYSEQSVAY CNSLRLYNSN VVFCI | 205 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 77 | 194 | 1.5e-27 | 0.8846153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10948 | CE4_BsPdaA_like | 4.12e-67 | 42 | 198 | 1 | 158 | Catalytic NodB homology domain of Bacillus subtilis polysaccharide deacetylase PdaA, and its bacterial homologs. The Bacillus subtilis genome contains six polysaccharide deacetylase gene homologs: pdaA, pdaB (previously known as ybaN), yheN, yjeA, yxkH and ylxY. This family is represented by Bacillus subtilis pdaA gene encoding polysaccharide deacetylase BsPdaA, which is a member of the carbohydrate esterase 4 (CE4) superfamily. BsPdaA deacetylates peptidoglycan N-acetylmuramic acid (MurNAc) residues to facilitate the formation of muramic delta-lactam, which is required for recognition of germination lytic enzymes. BsPdaA deficiency leads to the absence of muramic delta-lactam residues in the spore cortex. Like other CE4 esterases, BsPdaA consists of a single catalytic NodB homology domain that appears to adopt a deformed (beta/alpha)8 barrel fold with a putative substrate binding groove harboring the majority of the conserved residues. It utilizes a general acid/base catalytic mechanism involving a tetrahedral transition intermediate, where a water molecule functions as the nucleophile tightly associated to the zinc cofactor. |
| cd10917 | CE4_NodB_like_6s_7s | 1.75e-36 | 81 | 197 | 1 | 117 | Catalytic NodB homology domain of rhizobial NodB-like proteins. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes many rhizobial NodB chitooligosaccharide N-deacetylase (EC 3.5.1.-)-like proteins, mainly from bacteria and eukaryotes, such as chitin deacetylases (EC 3.5.1.41), bacterial peptidoglycan N-acetylglucosamine deacetylases (EC 3.5.1.-), and acetylxylan esterases (EC 3.1.1.72), which catalyze the N- or O-deacetylation of substrates such as acetylated chitin, peptidoglycan, and acetylated xylan. All members of this family contain a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold with 6- or 7 strands. Their catalytic activity is dependent on the presence of a divalent cation, preferably cobalt or zinc, and they employ a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine) to carry out acid/base catalysis. Several family members show diversity both in metal ion specificities and in the residues that coordinate the metal. |
| COG0726 | CDA1 | 1.45e-35 | 55 | 190 | 39 | 174 | Peptidoglycan/xylan/chitin deacetylase, PgdA/CDA1 family [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam01522 | Polysacc_deac_1 | 3.16e-34 | 76 | 194 | 2 | 119 | Polysaccharide deacetylase. This domain is found in polysaccharide deacetylase. This family of polysaccharide deacetylases includes NodB (nodulation protein B from Rhizobium) which is a chitooligosaccharide deacetylase. It also includes chitin deacetylase from yeast, and endoxylanases which hydrolyzes glucosidic bonds in xylan. |
| cd10954 | CE4_CtAXE_like | 2.29e-27 | 81 | 186 | 1 | 106 | Catalytic NodB homology domain of Clostridium thermocellum acetylxylan esterase and its bacterial homologs. This family is represented by Clostridium thermocellum acetylxylan esterase (CtAXE, EC 3.1.1.72), a member of the carbohydrate esterase 4 (CE4) superfamily. CtAXE deacetylates O-acetylated xylan, a key component of plant cell walls. It shows no detectable activity on generic esterase substrates including para-nitrophenyl acetate. It is specific for sugar-based substrates and will precipitate acetylxylan, as a consequence of deacetylation. CtAXE is a monomeric protein containing a catalytic NodB homology domain with the same overall topology and a deformed (beta/alpha)8 barrel fold as other CE4 esterases. However, due to differences in the topography of the substrate-binding groove, the chemistry of the active center, and metal ion coordination, CtAXE has different metal ion preference and lacks activity on N-acetyl substrates. It is significantly activated by Co2+. Moreover, CtAXE displays distinctly different ligand coordination to the metal ion, utilizing an aspartate, a histidine, and four water molecules, as opposed to the conserved His-His-Asp zinc-binding triad of other CE4 esterases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEG16570.1 | 1.09e-49 | 31 | 198 | 80 | 247 |
| AEM79381.1 | 3.99e-47 | 24 | 194 | 54 | 222 |
| AUG57418.1 | 4.17e-47 | 40 | 198 | 83 | 241 |
| ALX08146.1 | 7.77e-47 | 28 | 198 | 81 | 248 |
| ADU74203.1 | 7.77e-47 | 28 | 198 | 81 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2J13_A | 2.54e-38 | 46 | 198 | 20 | 173 | Structureof a family 4 carbohydrate esterase from Bacillus anthracis [Bacillus anthracis str. Ames] |
| 1W1A_1 | 3.27e-38 | 44 | 185 | 23 | 165 | Structureof Bacillus subtilis PdaA in complex with NAG, a family 4 Carbohydrate esterase. [Bacillus subtilis],1W1A_2 Structure of Bacillus subtilis PdaA in complex with NAG, a family 4 Carbohydrate esterase. [Bacillus subtilis],1W1B_1 Structure of Bacillus subtilis PdaA with Cadmium, a family 4 Carbohydrate esterase. [Bacillus subtilis],1W1B_2 Structure of Bacillus subtilis PdaA with Cadmium, a family 4 Carbohydrate esterase. [Bacillus subtilis] |
| 1W17_A | 3.80e-38 | 44 | 185 | 29 | 171 | Structureof Bacillus subtilis PdaA, a family 4 Carbohydrate esterase. [Bacillus subtilis],1W17_B Structure of Bacillus subtilis PdaA, a family 4 Carbohydrate esterase. [Bacillus subtilis] |
| 1NY1_A | 1.67e-37 | 44 | 185 | 6 | 148 | CrystalStructure Of B. Subtilis Polysaccharide Deacetylase Northeast Structural Genomics Consortium Target Sr127. [Bacillus subtilis],1NY1_B Crystal Structure Of B. Subtilis Polysaccharide Deacetylase Northeast Structural Genomics Consortium Target Sr127. [Bacillus subtilis] |
| 5O6Y_A | 3.79e-19 | 76 | 188 | 16 | 128 | Crystalstructure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q04729 | 1.83e-42 | 33 | 189 | 19 | 176 | Uncharacterized 30.6 kDa protein in fumA 3'region OS=Geobacillus stearothermophilus OX=1422 PE=3 SV=1 |
| O34928 | 2.08e-37 | 44 | 185 | 29 | 171 | Peptidoglycan-N-acetylmuramic acid deacetylase PdaA OS=Bacillus subtilis (strain 168) OX=224308 GN=pdaA PE=1 SV=1 |
| Q81EK9 | 5.75e-18 | 76 | 188 | 76 | 188 | Peptidoglycan-N-acetylglucosamine deacetylase BC_1960 OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=BC_1960 PE=1 SV=1 |
| Q52845 | 2.36e-17 | 81 | 186 | 21 | 127 | Chitooligosaccharide deacetylase OS=Mesorhizobium japonicum (strain LMG 29417 / CECT 9101 / MAFF 303099) OX=266835 GN=nodB PE=3 SV=2 |
| A0A0H3GDH9 | 6.82e-16 | 78 | 186 | 263 | 371 | Peptidoglycan-N-acetylglucosamine deacetylase PgdA OS=Listeria monocytogenes serotype 1/2a (strain 10403S) OX=393133 GN=pgdA PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
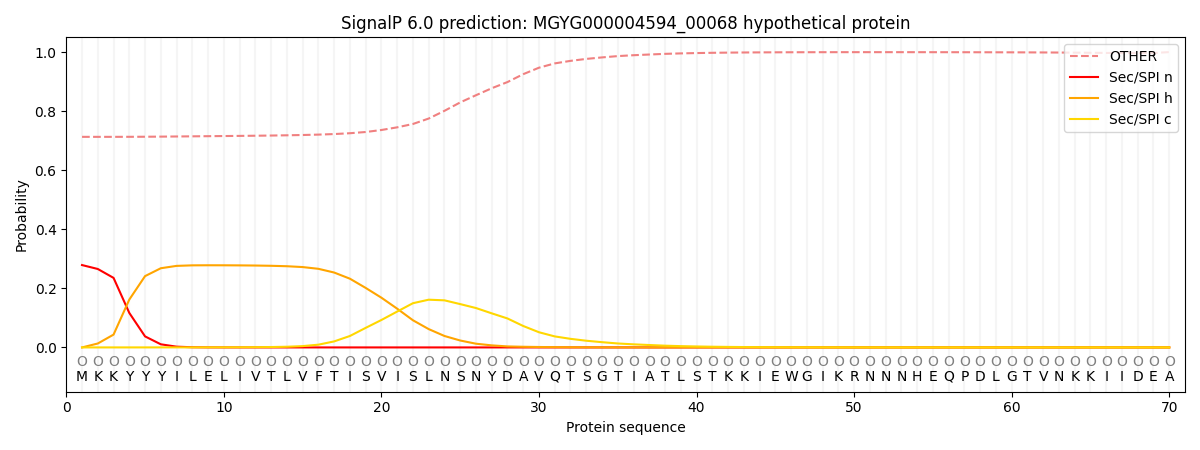
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.725982 | 0.262738 | 0.004891 | 0.000514 | 0.000396 | 0.005480 |