You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004627_00244
You are here: Home > Sequence: MGYG000004627_00244
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella nigrescens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella nigrescens | |||||||||||
| CAZyme ID | MGYG000004627_00244 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16488; End: 19895 Strand: + | |||||||||||
Full Sequence Download help
| MDRQVFQTNS RQRWNRFKWT LRVLLTIAML LGAVFLAMFA LEGSPRMPFR HDYRSVVSAG | 60 |
| KPFLKDNKTS KEYKSFRDFF QEQRMHSNYA RVATRQHRFV GKARGLAQKY IAEWTDPRMG | 120 |
| IRSAWYVNWD KRAYLSLKNN IKHLNMVLPE WFFIDPNTDQ LESRIDQKAL KLMHSAGVPI | 180 |
| LPMLTNNYGA KFRSEPIGRI MSDPKKRMAL INQLADQCKQ HGFVGINLDL EDLNINDNAL | 240 |
| LVTLVKDFAT VFHAHGLYVT QAIAAFNDDY DVQELAKYDD YLFLMAYDEH NSTSQPGAVS | 300 |
| SQRWVEKATD WAAKNVPNDK IVLGLAAYGY DWTEGKAVGT AVSFDQIIAT AQNAGAKIAF | 360 |
| NDDTYNLDFS YKDEGNGTLH HVFFPDAATT FNIMRFGTQY HMAGFGLWRL GTEDKRIWRF | 420 |
| YGKDMAWENA SKLSVSKLMQ LNGTDDVNFV GSGEVLNVTS EPHHGKIAVT IDKDNCLITE | 480 |
| EYYQALPTTY TIQRLGKCKP KQLVLTFDDG PDEHWTPTVL STLKKYKVPA AFFMVGLQME | 540 |
| KNLPLVKQVF DDGHTIGNHT FTHHDMSENS DRRSYAELKL TRMLIESITG QSTILFRAPY | 600 |
| NADADPTGHE EIWPMIIASR RNYLFVGESI DPNDWQPGVT ADQIYKRVID GVHNEDGHII | 660 |
| LLHDAGGDTR EATVAALPRI IETLQREGYQ FISLEQYLDM SRQTLMPPIE KGNVYYAMQA | 720 |
| NLSLAEFIYH VSDFLTALFL VFLVLGFVRL LFMYILMVRE KRTENHRNYA PISAGTAPEV | 780 |
| SIIVPAYNEE VNIVRTISNL KQQDYPSFHI YLVDDGSSDN TLARAREQFG NDAEVTIIEK | 840 |
| ENGGKASALN RGIAACTTEY VVCIDADTQL QSDAVSKLMR HFVADKTGRV GAVAGNVKVG | 900 |
| NQRNMLTYWQ AIEYTTSQNF DRMAYSNINA ITVIPGAIGA FRKKVLEEVG GFTTDTLAED | 960 |
| CDLTMSINER GYIIENENYA IALTEVPETL RQFVKQRIRW CFGVMQAFWK HRSSLFATSE | 1020 |
| KGFGMWAMPN MLVFQYIIPT FSPLADVLML IGLFTGNAAQ VFIYYLVFLL VDASVSIMAY | 1080 |
| IFEHERLWVL LWIIPQRFFY RWIMYYVLFK SYLKAIKGEL QTWGVLKRTG HVKSS | 1135 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 778 | 1003 | 8.5e-38 | 0.9956521739130435 |
| GH18 | 137 | 415 | 2.4e-34 | 0.847972972972973 |
| CE4 | 497 | 601 | 1.1e-28 | 0.7923076923076923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd10962 | CE4_GT2-like | 3.06e-96 | 501 | 699 | 1 | 196 | Catalytic NodB homology domain of uncharacterized bacterial glycosyl transferase, group 2-like family proteins. This family includes many uncharacterized bacterial proteins containing an N-terminal GH18 (glycosyl hydrolase, family 18) domain, a middle NodB-like homology domain, and a C-terminal GT2-like (glycosyl transferase group 2) domain. Although their biological function is unknown, members in this family contain a middle NodB homology domain that is similar to the catalytic domain of Streptococcus pneumoniae polysaccharide deacetylase PgdA (SpPgdA), an extracellular metal-dependent polysaccharide deacetylase with de-N-acetylase activity toward a hexamer of chitooligosaccharide N-acetylglucosamine, but not shorter chitooligosaccharides or a synthetic peptidoglycan tetrasaccharide. Like SpPgdA, this family is a member of the carbohydrate esterase 4 (CE4) superfamily. The presence of three domains suggests that members of this family may be multifunctional. |
| cd06423 | CESA_like | 4.09e-68 | 782 | 960 | 1 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| cd02874 | GH18_CFLE_spore_hydrolase | 5.51e-65 | 124 | 419 | 6 | 311 | Cortical fragment-lytic enzyme (CFLE) is a peptidoglycan hydrolase involved in bacterial endospore germination. CFLE is expressed as an inactive preprotein (called SleB) in the forespore compartment of sporulating cells. SleB translocates across the forespore inner membrane and is deposited as a mature enzyme in the cortex layer of the spore. As part of a sensory mechanism capable of initiating germination, CFLE degrades a spore-specific peptidoglycan constituent called muramic-acid delta-lactam that comprises the outer cortex. CFLE has a C-terminal glycosyl hydrolase family 18 (GH18) catalytic domain as well as two N-terminal LysM peptidoglycan-binding domains. In addition to SleB, this family includes YaaH, YdhD, and YvbX from Bacillus subtilis. |
| COG1215 | BcsA | 5.75e-64 | 727 | 1116 | 5 | 391 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| PRK11204 | PRK11204 | 3.42e-59 | 735 | 1123 | 1 | 411 | N-glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUB55043.1 | 0.0 | 1 | 1135 | 1 | 1135 |
| QUB50645.1 | 0.0 | 1 | 1135 | 1 | 1135 |
| QUB50174.1 | 0.0 | 1 | 1135 | 1 | 1135 |
| AXV48412.1 | 0.0 | 1 | 1134 | 1 | 1134 |
| QUB90351.1 | 0.0 | 1 | 1134 | 1 | 1134 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5O6Y_A | 9.67e-29 | 496 | 703 | 16 | 213 | Crystalstructure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
| 5O6Y_B | 1.13e-27 | 496 | 703 | 16 | 213 | Crystalstructure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579],5O6Y_C Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579],5O6Y_D Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide [Bacillus cereus ATCC 14579] |
| 7FBW_A | 1.54e-27 | 487 | 699 | 101 | 301 | ChainA, Predicted xylanase/chitin deacetylase [Caldanaerobacter subterraneus subsp. tengcongensis MB4] |
| 4L1G_A | 4.82e-27 | 496 | 703 | 68 | 265 | Crystalstructure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_B Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_C Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579],4L1G_D Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus [Bacillus cereus ATCC 14579] |
| 2C71_A | 5.55e-27 | 501 | 711 | 5 | 202 | ChainA, Glycoside Hydrolase, Family 11:clostridium Cellulosome Enzyme, Dockerin Type I:polysaccharide [Acetivibrio thermocellus],2C79_A Chain A, GLYCOSIDE HYDROLASE, FAMILY 11:CLOSTRIDIUM CELLULOSOME ENZYME, DOCKERIN TYPE I:POLYSACCHARIDE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XAR5 | 1.23e-33 | 754 | 1040 | 51 | 330 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Escherichia coli O157:H7 OX=83334 GN=pgaC PE=3 SV=1 |
| P75905 | 4.06e-33 | 754 | 1040 | 51 | 330 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Escherichia coli (strain K12) OX=83333 GN=pgaC PE=1 SV=1 |
| Q7A351 | 1.47e-32 | 754 | 1003 | 24 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain N315) OX=158879 GN=icaA PE=3 SV=1 |
| Q8GLC5 | 1.47e-32 | 780 | 1009 | 49 | 280 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus epidermidis OX=1282 GN=icaA PE=3 SV=1 |
| Q6GDD8 | 1.47e-32 | 754 | 1003 | 24 | 270 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=icaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000048 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
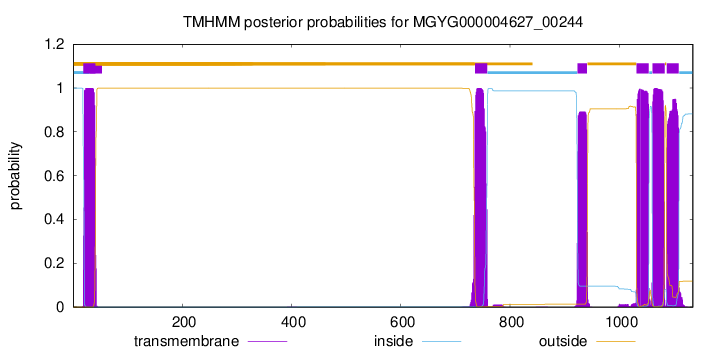
| start | end |
|---|---|
| 19 | 41 |
| 736 | 758 |
| 924 | 941 |
| 1032 | 1054 |
| 1061 | 1083 |
| 1087 | 1109 |
