You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000346_00162
You are here: Home > Sequence: MGYG000000346_00162
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
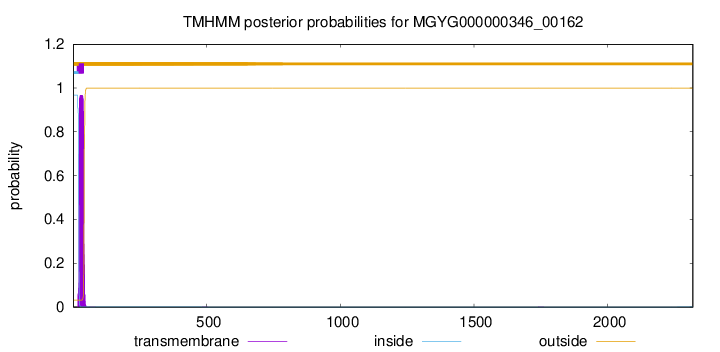
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000346_00162 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2883; End: 9845 Strand: - | |||||||||||
Full Sequence Download help
| MTYKEVKQKK GHKHLKAGLK RILACVLSSA LLFSGMPIGI DFSQGVISAK AAVDIDDLTG | 60 |
| TAGSEAVSGP GAAILAAPAA FGMERAAAQT TAWTPGNSEL FDASDTSVTE IAPGSTGTLT | 120 |
| NSDGYSFLIN SGKFAVRTGN GDFQVNAGAV IEIPIISGAK SCTIELTAYS SVSDTATFEG | 180 |
| LTDINITESG SGWKTYTVSG RPEDSVSTVT VSLTGNNYFR SIGVSSSSSS ATTSALLADD | 240 |
| HMQASWGYTD TLLDSSGAAA TQLQKSTGTY TNAEGDVLYV DATAGKFAPT AEKTRIQVNN | 300 |
| GTKIYVPVKG SKTTIEVSMN KNCVTTADAL LSDVLTVNGI NNNLVSAKCI SLAADAADSN | 360 |
| YRMATIECYT NGEEGEILLT SAYADSNYFR SISVSCEELN AVNISGTVTA EEAITGNLQV | 420 |
| IILNTATNAQ YAAEVTNGRY SVDVLVEAAD AVYEISINSQ EYQVSAGTTQ VTLNNASANV | 480 |
| ISNLTLAKMS TYILSGNILG IASGYDVSNL ELVFTAEANS AFIPVVSVNT EQMTYTAKLE | 540 |
| KGVSYSVELL GVNDYEPEQK MTGINHTADS NMDISVVMKS RYPITLSLGS TPDLSSMSVV | 600 |
| YNFTHGDGSV YKFTDKSAIV LRDGAYTVSI GGEFEQQAYK ISLGTTLTVS GQAVNHAIAF | 660 |
| SPVTSWSFAV GSGDYYNKTI QNGTGYYQGL YIDASAGKLV PNGAVETANS AQFNTGAVIH | 720 |
| VPVTGKCSIE VKAYQAQYAY YTINGTAADT ASASTTIEYD GGAGYVDIVS TGSAYLVGIT | 780 |
| VIYPAKDVQY VEQPVMPFVA GDDTDANTAY DTDQIPRASK SESIVVQPVG QKLNITQTGG | 840 |
| EITGAYSKIS NLGYYLFPMT TDNNRLEFDM VIHSSGSSNG DGAYMGLFTN NYAYTLGIRK | 900 |
| GTNARAIYSK AAVNQGDYAG AGGINETLEI GTPLHIEISL QSGKPAASYT IIETGETASV | 960 |
| SLSTLADEPG YYYGLVVSDA SVTITNMRYT AEDGTILYDQ NACYYPEGTM PAVISVTAQA | 1020 |
| ADSREYIDVS WTGSVPEGDG TYVVQMKKDN GDWADLADDV TGFSYRYMLP EGEGGNYLFR | 1080 |
| VCGQLGKAAL GGSRNTFVEM TEAVYVLSAL AKPVVTAKAD ASGIALDWQD VASAEYYQIY | 1140 |
| RYSFDEGADA VKCIATVGES AYKDTAAQAE MPYYYSVKAV SDTEANFSPF SDIIWAVATA | 1200 |
| GHTGEYVYED EATDITITKK SYDTVFTDKI VLEGIVAGAG TLTAYVNGKS AAEQDMSAGE | 1260 |
| SFSFELTAEE GRNDISLIFT DTNGDKTRKT FNFVYLTNYD IVADASYAGT DGELVNGIPT | 1320 |
| YKTVQAAVDS VPAGNSDTKV ILVMAGDYEE RLVVNTPNIS LVGEDRESTL IHYYPGVLGS | 1380 |
| AYEAGGDMDK RCAVYIQSGA ESFSAENISF ANDYVYSTPD SKSNKSADAL RCDAEGVVFV | 1440 |
| NVKLSGVQDT LYMHQGHQYY YKCRIEGLID FIYSGDNARA FFNDCEIVFV YESTKTSGYV | 1500 |
| CAPKTAEDAD YGLTFYDCVV IGEEGCSGTG YLLARPWGAS AYITWISCYM GRSVYKIMPY | 1560 |
| GAMSGNQPEN ARFFEFGTYG PGYAINADRR QISPNKAEEM ISDSYLGWSP ASEADTVASH | 1620 |
| YIGSIVTDRG PMYVETEASS DTYLWTDGDD TGLKAYDLEG YAEGYGVSGG GLLKENNDNY | 1680 |
| YQVGTAEEFL NALIAIKSSG KKSVIELTAD INLGCNEIAD FDSYSSIIKP YSAQALTHPT | 1740 |
| LIASGVSVLN FTNIYNLTIF SLNGSSIKHA NITMKNSENI IIRNIKFDEL WEWDEATSGD | 1800 |
| YDRNDWDYMT IDQGCNGIWI DHCTFYKAYD GVIDVKNPAP EENVTISWCE FLPGSEDNIF | 1860 |
| FDVMMNELAA DPAAYPAYQH MLDEGMTAGQ IYMYAYGQKK THLFGQSDEA TNAAGIQVTL | 1920 |
| ANNYYKNSMD RMPRLRYGYS HVYNCIMDAQ DLLDAKMTIT DPDIAKKIVS NGASSTCGAQ | 1980 |
| VLLENCYING ILNALNSGNG SSPSGYINAI NSVYYMDGEL AELEPKCNST GDTRVLVTDA | 2040 |
| DAFISALPYS DYVLYDAEQL SAVVMPYAGA GKLDLTVLQW EKASYNAVLE IPDVPEEPEE | 2100 |
| PDEPDTPDES DDDDYDSDDS YTSSSSSSAS ASNNANIVQK PDAEDESGSG TWKKNDKGWW | 2160 |
| FEYSADGSYA VNSWHKIGGE WYSFDEEGYM ETGWKYDKSY ESWFYLEDSG AMATSWVYDE | 2220 |
| SYGSWFYMED NGAMATDWIY DESYGSWFYL DGSGAMKTGW LFDARYDGWF YLEDSGAMKV | 2280 |
| GWLYDAARNV WYYLKSNGIM AVNEMTPDGY RVNADGIWTE | 2320 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1318 | 1605 | 2.6e-67 | 0.9583333333333334 |
| PL1 | 1751 | 1988 | 5.8e-40 | 0.8168316831683168 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5263 | COG5263 | 8.78e-43 | 2154 | 2318 | 155 | 312 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033838 | PspC_subgroup_1 | 1.11e-39 | 2139 | 2318 | 476 | 682 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| NF033930 | pneumo_PspA | 7.10e-39 | 2139 | 2316 | 432 | 599 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| PLN02773 | PLN02773 | 4.03e-37 | 1321 | 1606 | 17 | 296 | pectinesterase |
| COG4677 | PemB | 1.00e-34 | 1308 | 1599 | 87 | 395 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADD62022.1 | 0.0 | 84 | 2086 | 25 | 2030 |
| QEH70392.1 | 0.0 | 819 | 2086 | 497 | 1754 |
| ACZ98653.1 | 0.0 | 808 | 2085 | 74 | 1348 |
| ACR72234.1 | 2.91e-117 | 830 | 1612 | 279 | 1071 |
| AWV33413.1 | 1.81e-83 | 1653 | 2086 | 108 | 550 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 3.80e-77 | 1656 | 2085 | 7 | 446 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
| 2NSP_A | 1.17e-33 | 1297 | 1610 | 2 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 2.52e-29 | 1297 | 1610 | 2 | 339 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 8.43e-29 | 1297 | 1610 | 2 | 339 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 3UW0_A | 1.03e-24 | 1321 | 1593 | 44 | 345 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 3.57e-72 | 1656 | 1988 | 36 | 370 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
| P0C1A8 | 3.00e-28 | 1297 | 1610 | 26 | 363 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| P0C1A9 | 7.39e-28 | 1297 | 1610 | 26 | 363 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| Q9LVQ0 | 1.44e-23 | 1316 | 1606 | 12 | 296 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| Q9FM79 | 2.56e-22 | 1316 | 1579 | 87 | 339 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
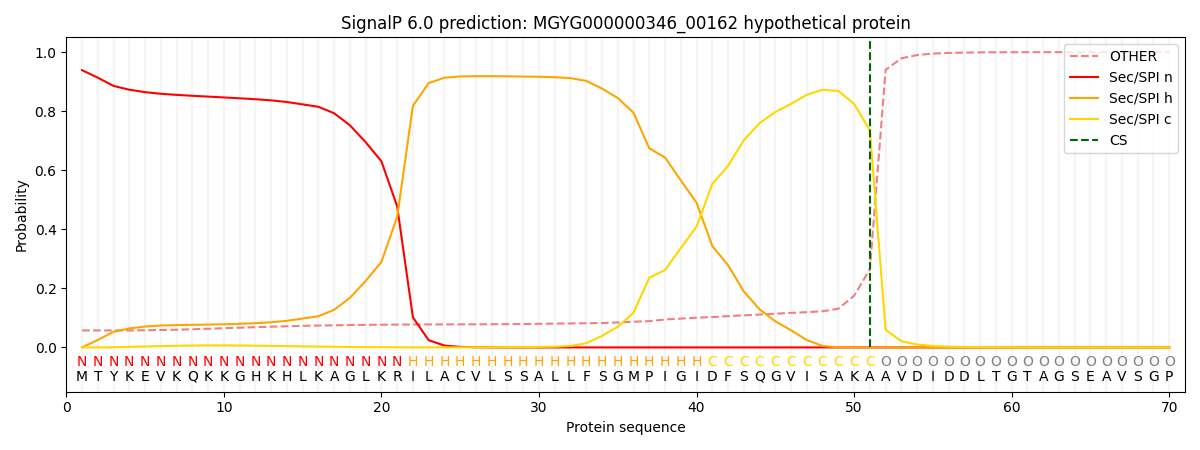
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.060209 | 0.934432 | 0.004431 | 0.000387 | 0.000264 | 0.000262 |