You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000562_00359
You are here: Home > Sequence: MGYG000000562_00359
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
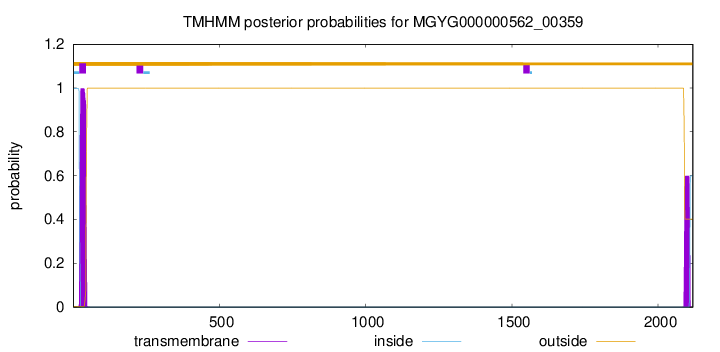
TMHMM annotations
Basic Information help
| Species | Lachnospira sp000437735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira sp000437735 | |||||||||||
| CAZyme ID | MGYG000000562_00359 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3115; End: 9474 Strand: + | |||||||||||
Full Sequence Download help
| MNQKTQNRAQ RRRANRHHVK RATAFALAGL MATSGISLPA GTFAYGAENN TLLAFAGAEG | 60 |
| GGRFATGGRG YDVYVVTTLE DYGKGETPVE GSLRYGIEEF AKDKGGTMIV FNVGGTIELK | 120 |
| QTLTFKERKN ITVAGQTAPG DGITLSGWDT NISNSENLII RYMRFRPGAA NVHTGGDSMD | 180 |
| ALWGRDNKTF MIDHCSFSWN TDETVSTYRG QDGTVQWCIV SESLTVSGHS KGRHGYGGIF | 240 |
| GGDNVLFQNN LIANHTSRNP RIGGGCMGDP TKDGGSTATL QLSNNVLYNW GYNTCYGGGY | 300 |
| AYTNFINNFL KPGQGTREQV RYQVIDMGEA TKPGGFYVNG NYMDGNAEIT ADNAKGSKMS | 360 |
| GVTEGANKTV VSETPYTAEG FDSATVTSAT DCYEPVLAQA GATYPYRDAI DARVVAETRT | 420 |
| DSGRYVNTED EVGGYPAKES VRAASFDTDM DGIPDTWETA HGLNPNDASD SKKINSATGY | 480 |
| AYVEEYFNGL VENVEQSGYT APNPDITLDL QDNAQYDEGQ TVTVTADVKA NNGGSIAKVE | 540 |
| FYNGIQLVGT ADTAPFTFDY TGLTDGTYNI TARAYDNDGN QTQSSVSKLH INSTAGTGEW | 600 |
| TSTDIGNPGV AGNASLENGV MTVKGAGKLG ASEGCVSGST YADATTDDFQ YTYQLVKGDA | 660 |
| EIITKLDSAT TVDNHVFTGV MFRESLESGS KTAALGMSMV KISNETTWST YLASRLETNG | 720 |
| KISDISETID SPASAEKAGI PLVRDLHFKS GADFNGTWFK LIRRGDTFTG YASDDGVTWT | 780 |
| KVGSKTIEMA QDIYVGFAVD ANKAANSLEN LSTAKFSNIA IHEEFTDVDY NLEHITTSGA | 840 |
| DYAAVGTDFT TQLTADSGYH LPDAIEIKAG ENVLAKQDYT YDAKTGDIVV KADRLTSGVK | 900 |
| LTISAQAEED VKIDYTLQTY GDTEDVTVTQ DAGSMTISQT AESASMVGNK GTAGKNVSYI | 960 |
| LFPQSKEAAK MTLKLKVNSF SNGKYSGVYA GAFQPDGDYL YNSIGFRGTG DVNAVSPYWL | 1020 |
| KASGKAGNGS PKQAWTEGAV YTVSIEKKAD NTYYATFQEE GSAAVNEKTF KASDSALTPD | 1080 |
| IMAQFGLAVY SADVTVTDMV LSDMSGNVLY DQNRDNSGQE ESGYKPQVSG SDLLTVTGAG | 1140 |
| SVMKAEQSAE SGSITQSTGD AGVNTSYILL PENDAFASMS MTLKITDYSI TGQNAKRTGV | 1200 |
| FVGAFQTSGK YAFASLGFRG YDSSVGTDAL SGYWMKGTGK AGNGSPKYEV AADNKYNVTF | 1260 |
| TRTENGYLAE FESLGKPLLV DANGNKTYSD KKEFKWKDMT LTAEDAVSYG LALVGADVEI | 1320 |
| TNWVAKDAEG NILYNQKDYY KDAGTAPAVT GVSTPVISED RSSITVSWSG EGAQLDGKYR | 1380 |
| VEVSQDNGVT YKELDTTAET SYSYVPENSG DYQFRITGVC GGQESNSMET SSVHFVLPMT | 1440 |
| APSIGAESGN AANTVTWSAV PEAVSYRIYR STQRAEGYTE IGTSESTTFT DTTAENEVPY | 1500 |
| FYTVAAVGQD NESNPSAPVS IMATAGHTGE YQFGSQAAQM TVLEKSNDTV TENEAALKFA | 1560 |
| YDEAGKVSVY AGDRLLSEKE TAAGEAVDIT IPLQDGRNAV KVVFENTQGI KTYRNFNFVK | 1620 |
| LSSYDMVVDA SGIRTLDSES VPVYTTVAEA LAAVPADNQE QKVIFVKNGT YYEKLSITSP | 1680 |
| YITLIGEDSE KTVLCYDAAS GKTDPVTGAA YGTSGSASVS IEAAAHHTYM ENLTVANTFD | 1740 |
| YPNETIEGKQ AVAMLTRADQ LIFNNVRLTG WQDTLQADGG GRQYFKNCYI EGNVDWIFGS | 1800 |
| AQAVFDDCDI VANAAGYVTA ASTESTKTTG YVFINSRLLK KSDSVAENSV ALGRPWRSNA | 1860 |
| CVTYVKCFMD SHIKTKGYDN MSGNTHSAAR FYEYQSYGPG FAVNTDRRQL AKAEGESLTV | 1920 |
| NGVFAREAGE GMAFAEGWDA VAAYAAASAD YTESGAVSVD FSELDRAIQN AEGLNSADYK | 1980 |
| DFSTVESALN AAKALDRTTA TQEEVSVLAH RLIEAVANLE TMTPAPEPTP DPTPTPEPTP | 2040 |
| TPTPTPSEPD NNQPGDTGEA DGSQNGNDNP SENAAENQQE SVQTADKTHS VWYLFGAAVS | 2100 |
| AMLAGFAGSK RKKKSDEEL | 2119 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1643 | 1922 | 7.1e-84 | 0.9479166666666666 |
| PL1 | 114 | 309 | 3.5e-65 | 0.9890710382513661 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 5.74e-50 | 1644 | 1924 | 94 | 402 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02773 | PLN02773 | 2.72e-48 | 1644 | 1918 | 17 | 290 | pectinesterase |
| PLN02682 | PLN02682 | 5.75e-42 | 1644 | 1900 | 82 | 334 | pectinesterase family protein |
| pfam01095 | Pectinesterase | 6.52e-42 | 1644 | 1908 | 12 | 271 | Pectinesterase. |
| PLN02665 | PLN02665 | 2.50e-38 | 1644 | 1914 | 80 | 344 | pectinesterase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYR21100.1 | 2.39e-254 | 20 | 821 | 9 | 832 |
| AWV33235.1 | 7.99e-246 | 51 | 828 | 52 | 846 |
| AIQ73887.1 | 9.62e-245 | 51 | 828 | 46 | 840 |
| AOZ92378.1 | 1.66e-229 | 48 | 1920 | 343 | 2132 |
| QNF31079.1 | 2.03e-196 | 47 | 829 | 23 | 756 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 1.30e-47 | 1644 | 1924 | 44 | 358 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 2NSP_A | 4.48e-45 | 1622 | 1919 | 2 | 329 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 4.86e-40 | 1622 | 1919 | 2 | 329 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 1.64e-39 | 1622 | 1919 | 2 | 329 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 5C1E_A | 1.10e-32 | 1631 | 1900 | 8 | 265 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5B297 | 2.53e-47 | 48 | 491 | 16 | 411 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 4.18e-45 | 50 | 491 | 18 | 414 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| B8NQQ7 | 1.40e-44 | 50 | 491 | 18 | 414 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| A1DPF0 | 1.44e-44 | 52 | 491 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| B0XMA2 | 6.52e-44 | 52 | 491 | 21 | 415 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
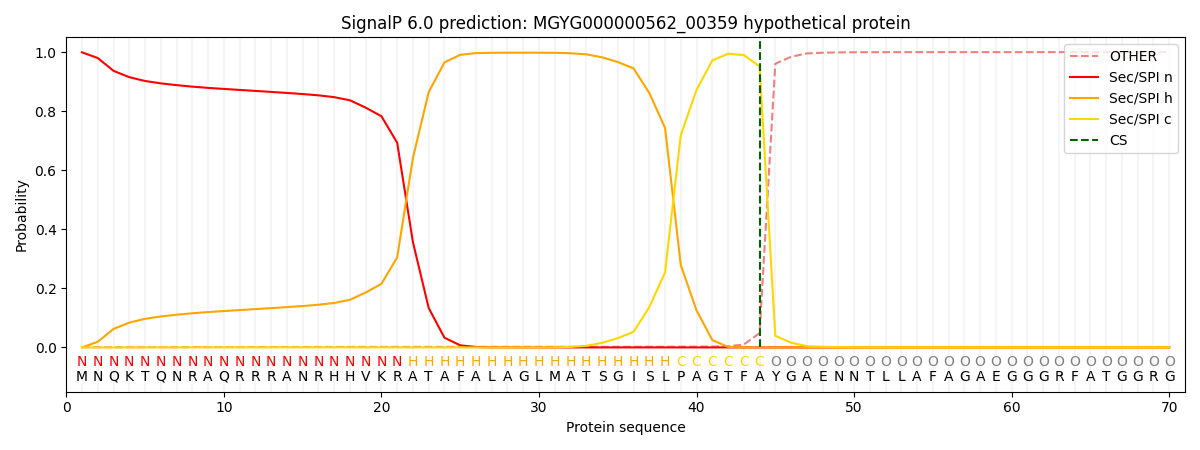
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001725 | 0.997023 | 0.000370 | 0.000434 | 0.000226 | 0.000186 |