You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000003540_00288
You are here: Home > Sequence: MGYG000003540_00288
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1232 sp900769385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; UBA1232; UBA1232 sp900769385 | |||||||||||
| CAZyme ID | MGYG000003540_00288 | |||||||||||
| CAZy Family | CE9 | |||||||||||
| CAZyme Description | N-acetylglucosamine-6-phosphate deacetylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2908; End: 4065 Strand: + | |||||||||||
Full Sequence Download help
| MLTQIINGKI HTPQGWLKNG SVIISDNKIL EVSNCDLAVV GAKIVDARGM YIVPGGVEIH | 60 |
| VHGGGGRDFM EGTEEAFRTA VDAHMQHGTT SIFPTLSSSS IPMIRAAAAT TEKLMAEKDS | 120 |
| PILGLHLEGH YFNMKMAGGQ IPEYIKNPDP NEYIPLLEET NCIKRWDASP ELPGALQFGK | 180 |
| YVSNKGILVS VAHTQAEFED IKAGFDVGYT HATHFYNAMP GFHKKNEYKY EGTVESIYLM | 240 |
| DDMTVEAIAD GIHVPPTILR LIYKVKGVEK TALITDALAC AASDSQTAFD PRVIIEDGVC | 300 |
| KLADRSALAG SIATMDRLIQ TMTLKAEVPL FDVCRMISET PAKIMGVYDR KGSLERGKDA | 360 |
| DILLLDNNLD VKAVWAMGNL VKNFE | 385 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE9 | 6 | 378 | 7.9e-105 | 0.9946380697050938 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00854 | NagA | 1.62e-136 | 5 | 378 | 2 | 374 | N-acetylglucosamine-6-phosphate deacetylase, NagA, catalyzes the hydrolysis of the N-acetyl group of N-acetyl-glucosamine-6-phosphate (GlcNAc-6-P) to glucosamine 6-phosphate and acetate. This is the first committed step in the biosynthetic pathway to amino-sugar-nucleotides, which is needed for cell wall peptidoglycan and teichoic acid biosynthesis. Deacetylation of N-acetylglucosamine is also important in lipopolysaccharide synthesis and cell wall recycling. |
| COG1820 | NagA | 5.44e-109 | 5 | 381 | 4 | 378 | N-acetylglucosamine-6-phosphate deacetylase [Carbohydrate transport and metabolism]. |
| TIGR00221 | nagA | 3.42e-81 | 5 | 379 | 7 | 380 | N-acetylglucosamine-6-phosphate deacetylase. [Central intermediary metabolism, Amino sugars] |
| PRK11170 | nagA | 2.94e-56 | 5 | 383 | 4 | 381 | N-acetylglucosamine-6-phosphate deacetylase; Provisional |
| pfam01979 | Amidohydro_1 | 1.70e-22 | 51 | 381 | 1 | 335 | Amidohydrolase family. This family of enzymes are a a large metal dependent hydrolase superfamily. The family includes Adenine deaminase EC:3.5.4.2 that hydrolyzes adenine to form hypoxanthine and ammonia. Adenine deaminases reaction is important for adenine utilisation as a purine and also as a nitrogen source. This family also includes dihydroorotase and N-acetylglucosamine-6-phosphate deacetylases, EC:3.5.1.25 These enzymes catalyze the reaction N-acetyl-D-glucosamine 6-phosphate + H2O <=> D-glucosamine 6-phosphate + acetate. This family includes the catalytic domain of urease alpha subunit. Dihydroorotases (EC:3.5.2.3) are also included. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49240.1 | 3.50e-242 | 1 | 382 | 1 | 382 |
| QIX66124.1 | 3.90e-239 | 1 | 382 | 1 | 382 |
| QCY55374.1 | 5.54e-239 | 1 | 382 | 1 | 382 |
| QRO17768.1 | 5.54e-239 | 1 | 382 | 1 | 382 |
| BBK89953.1 | 5.54e-239 | 1 | 382 | 1 | 382 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EGJ_A | 1.81e-44 | 5 | 383 | 7 | 381 | N-acetylglucosamine-6-phosphatedeacetylase from Vibrio cholerae. [Vibrio cholerae],3EGJ_B N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae. [Vibrio cholerae],3IV8_A N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate [Vibrio cholerae],3IV8_B N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate [Vibrio cholerae],3IV8_C N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate [Vibrio cholerae],3IV8_D N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate [Vibrio cholerae] |
| 2VHL_A | 5.13e-42 | 10 | 380 | 12 | 386 | TheThree-dimensional structure of the N-Acetylglucosamine-6- phosphate deacetylase from Bacillus subtilis [Bacillus subtilis],2VHL_B The Three-dimensional structure of the N-Acetylglucosamine-6- phosphate deacetylase from Bacillus subtilis [Bacillus subtilis] |
| 6FV3_A | 8.03e-41 | 30 | 381 | 36 | 392 | Crystalstructure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. [Mycolicibacterium smegmatis MC2 155],6FV3_B Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. [Mycolicibacterium smegmatis MC2 155],6FV3_C Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. [Mycolicibacterium smegmatis MC2 155],6FV3_D Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. [Mycolicibacterium smegmatis MC2 155] |
| 6FV4_A | 1.13e-39 | 30 | 381 | 36 | 392 | Thestructure of N-acetyl-D-glucosamine-6-phosphate deacetylase D267A mutant from Mycobacterium smegmatis in complex with N-acetyl-D-glucosamine-6-phosphate [Mycolicibacterium smegmatis MC2 155],6FV4_B The structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D267A mutant from Mycobacterium smegmatis in complex with N-acetyl-D-glucosamine-6-phosphate [Mycolicibacterium smegmatis MC2 155] |
| 1YMY_A | 1.51e-39 | 5 | 381 | 4 | 379 | CrystalStructure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 [Escherichia coli K-12],1YMY_B Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 [Escherichia coli K-12],1YRR_A Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution [Escherichia coli],1YRR_B Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution [Escherichia coli],2P50_A Crystal structure of N-acetyl-D-Glucosamine-6-Phosphate deacetylase liganded with Zn [Escherichia coli K-12],2P50_B Crystal structure of N-acetyl-D-Glucosamine-6-Phosphate deacetylase liganded with Zn [Escherichia coli K-12],2P50_C Crystal structure of N-acetyl-D-Glucosamine-6-Phosphate deacetylase liganded with Zn [Escherichia coli K-12],2P50_D Crystal structure of N-acetyl-D-Glucosamine-6-Phosphate deacetylase liganded with Zn [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O32445 | 9.30e-44 | 5 | 383 | 4 | 378 | N-acetylglucosamine-6-phosphate deacetylase OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=nagA PE=1 SV=2 |
| O34450 | 2.81e-41 | 10 | 380 | 12 | 386 | N-acetylglucosamine-6-phosphate deacetylase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagA PE=1 SV=1 |
| P44537 | 1.11e-39 | 5 | 383 | 5 | 381 | N-acetylglucosamine-6-phosphate deacetylase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=nagA PE=3 SV=1 |
| P0AF18 | 8.26e-39 | 5 | 381 | 4 | 379 | N-acetylglucosamine-6-phosphate deacetylase OS=Escherichia coli (strain K12) OX=83333 GN=nagA PE=1 SV=1 |
| P0AF19 | 8.26e-39 | 5 | 381 | 4 | 379 | N-acetylglucosamine-6-phosphate deacetylase OS=Escherichia coli O157:H7 OX=83334 GN=nagA PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
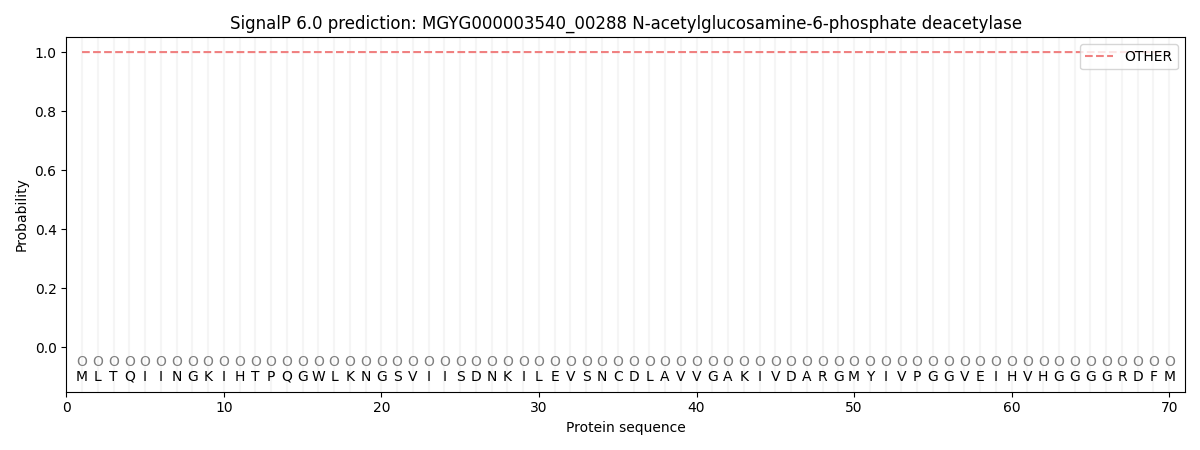
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000095 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
