You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001418_00684
You are here: Home > Sequence: MGYG000001418_00684
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromicrobium massiliense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; Aeromicrobium massiliense | |||||||||||
| CAZyme ID | MGYG000001418_00684 | |||||||||||
| CAZy Family | GH0 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 670514; End: 672982 Strand: - | |||||||||||
Full Sequence Download help
| MTDRSPATVR LLLTAVLGLL LTLVGGPAQA APEGPDEGRA WFGPNLDWEK QTATGYAERL | 60 |
| GATPSLYAQR VHYPLTDDDV TYLDQFVQQA TSQGAVAVVE LQPQVLLDQL SEQDAQRLAG | 120 |
| RLDALERERD ADVLVRFAPE MNASWVTWGQ SPADYRRAFR VVADAVHAEA PNAAMVWSPA | 180 |
| YGAGYPYGTA YGALETVDPE RSAALDTDDD GDVTEADDPY GPYYPGDAAV DWVGLSLYHF | 240 |
| GEVQDFGENV RPDDGDFAAR LDEQAGYGDE RRRRAFYERF AEGRDQPMLV ETGALFNTES | 300 |
| RGGDDELDVK RTWWRQVLAE LPSHPRIAGV TWLELRRQEA EADGDVVDWR AVHRDELADD | 360 |
| LRADLEDSSV ELGPVTRVLD QETGNEASQQ GWQPDDVSAE MGWIVLCVVL LAVAYLFSGL | 420 |
| AGRYLRSWRY PDEDDPRDRR LDLFRGWIIL AVVITHIEVS GPYSYVTLNA IGAITGAEMF | 480 |
| VLLSGVVLGM IYAPTVKRFG EWATAVTMWR RARKQYLVAL AVVMIVYLLS LLPFVDASVI | 540 |
| TTFTDRGTGA EGEGASGTVY DLYANAPRLF DYPPPWYAVK QLLLLQIGPW VFNIMGLFVV | 600 |
| LSLLLPVIMW FVRRGLWWLV LAVSWGLYLL DAAYGVRVLP SQFQDVFPLL TWQLPFVHGL | 660 |
| VLGHYRRPIQ RALTTRLGKV LVGVVVVGYA AALGYLWLCH RFEVRPDLLP ANAYGYLYDH | 720 |
| AYIRVFLQWG RLLDLAVMVV VAYAFLTTCW KPVNAAIGWL WIPLGQASLY VFIVHVFFVL | 780 |
| AVGNIPGLDR MSIWQGALVH TAVILTIWVM VKKRFLFSVI PR | 822 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam10129 | OpgC_C | 1.76e-72 | 437 | 818 | 1 | 357 | OpgC protein. This domain, found in various hypothetical and OpgC prokaryotic proteins. It is likely to act as an acyltransferase enzyme. |
| COG4645 | OpgC | 1.45e-15 | 418 | 667 | 4 | 233 | Predicted acyltransferase [General function prediction only]. |
| pfam01757 | Acyl_transf_3 | 4.25e-08 | 441 | 811 | 4 | 326 | Acyltransferase family. This family includes a range of acyltransferase enzymes. This domain is found in many as yet uncharacterized C. elegans proteins and it is approximately 300 amino acids long. |
| COG1835 | OafA | 6.51e-05 | 437 | 807 | 14 | 343 | Peptidoglycan/LPS O-acetylase OafA/YrhL, contains acyltransferase and SGNH-hydrolase domains [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVT80157.1 | 0.0 | 2 | 822 | 11 | 841 |
| QIG45368.1 | 0.0 | 13 | 822 | 12 | 846 |
| QNG36630.1 | 1.65e-310 | 12 | 821 | 6 | 820 |
| QOD43196.1 | 1.72e-308 | 31 | 821 | 64 | 873 |
| ALD14240.1 | 4.08e-302 | 6 | 821 | 5 | 844 |
SignalP and Lipop Annotations help
This protein is predicted as SP
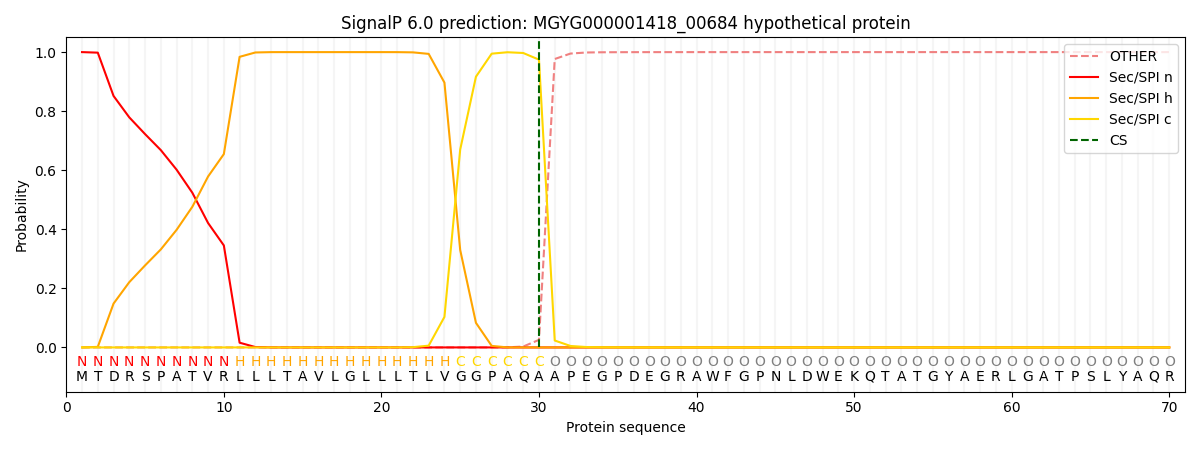
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.998905 | 0.000275 | 0.000180 | 0.000163 | 0.000148 |
TMHMM Annotations download full data without filtering help
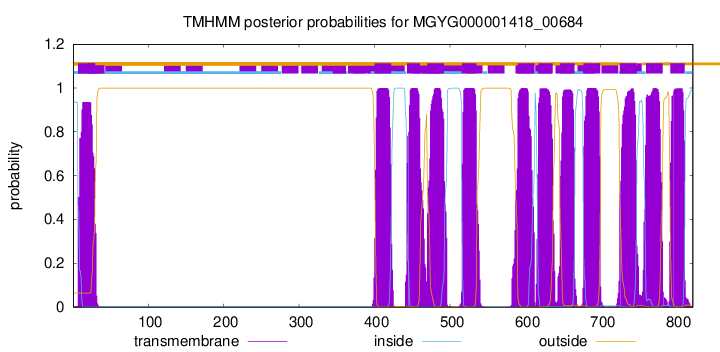
| start | end |
|---|---|
| 7 | 29 |
| 400 | 422 |
| 443 | 460 |
| 470 | 492 |
| 516 | 535 |
| 590 | 612 |
| 617 | 636 |
| 646 | 665 |
| 677 | 699 |
| 725 | 747 |
| 760 | 782 |
| 792 | 811 |
