You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001784_01341
You are here: Home > Sequence: MGYG000001784_01341
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mailhella sp003150275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Mailhella; Mailhella sp003150275 | |||||||||||
| CAZyme ID | MGYG000001784_01341 | |||||||||||
| CAZy Family | GH103 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11781; End: 12881 Strand: + | |||||||||||
Full Sequence Download help
| MDCSMKGEDM TRGSWFRLFL ILPLCLAAGS CSRAVDTGRP AEAALPPPAA EETAGGTVTL | 60 |
| ENVPEVWRPL VRQLEEDGIA GQEISWLFVR LGDELSQEPM GHKVRELYNV KYLPKPGSRE | 120 |
| QAPAVEGGDP IPKPWYKGVV TDANARRCRS FINMYAPAFH TAEETYGVPT EVAAALLFVE | 180 |
| TRLGDYLGDQ NAFVTLASMA ASRSPESIPD WMAALPDQAP EKLEWISRRM ADKADWAYAE | 240 |
| LRHLLQYSLK NGVDPFEIPS SRYGAVGLCQ FMPSSVARFA QDGNGDGIID LFDPADAVAS | 300 |
| LSNYLALHGW KRDSTIPQKI KVLRRYNNLA IYANTILALS EKIREIPGRP AVNRPLPKPS | 360 |
| AVVSQR | 366 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH103 | 134 | 321 | 3.3e-42 | 0.5423728813559322 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13406 | SLT_2 | 4.87e-45 | 133 | 353 | 48 | 248 | Transglycosylase SLT domain. This family is related to the SLT domain pfam01464. |
| PRK10760 | PRK10760 | 1.00e-28 | 119 | 316 | 96 | 269 | murein hydrolase B; Provisional |
| COG2951 | MltB | 2.78e-27 | 123 | 337 | 73 | 269 | Membrane-bound lytic murein transglycosylase B [Cell wall/membrane/envelope biogenesis]. |
| cd13399 | Slt35-like | 4.56e-25 | 260 | 342 | 24 | 108 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam01464 | SLT | 3.34e-05 | 246 | 306 | 3 | 75 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADP85790.1 | 1.73e-98 | 21 | 362 | 14 | 348 |
| AAS95158.1 | 1.73e-98 | 21 | 362 | 14 | 348 |
| ABM29301.1 | 6.96e-98 | 21 | 362 | 14 | 348 |
| SFV73295.1 | 8.20e-90 | 28 | 343 | 55 | 362 |
| QTO41794.1 | 1.17e-85 | 67 | 347 | 148 | 419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1LTM_A | 6.02e-20 | 137 | 316 | 78 | 230 | AcceleratedX-ray Structure Elucidation Of A 36 Kda Muramidase/transglycosylase Using Warp [Escherichia coli] |
| 1D0K_A | 6.18e-20 | 137 | 316 | 80 | 232 | ChainA, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1D0L_A Chain A, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1D0M_A Chain A, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE [Escherichia coli],1QDR_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QDT_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QUS_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli],1QUT_A Chain A, LYTIC MUREIN TRANSGLYCOSYLASE B [Escherichia coli] |
| 5O8X_A | 6.06e-19 | 140 | 310 | 61 | 205 | TheX-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa],5O8X_B The X-ray Structure of Catenated Lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 4ANR_A | 7.52e-19 | 140 | 310 | 78 | 222 | Crystalstructure of soluble lytic Transglycosylase SltB1 from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5ANZ_A | 1.06e-08 | 135 | 310 | 99 | 247 | CrystalStructure of SltB3 from Pseudomonas aeruginosa. [Pseudomonas aeruginosa],5AO7_A Crystal Structure of SltB3 from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide [Pseudomonas aeruginosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P41052 | 5.29e-19 | 137 | 316 | 119 | 271 | Membrane-bound lytic murein transglycosylase B OS=Escherichia coli (strain K12) OX=83333 GN=mltB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
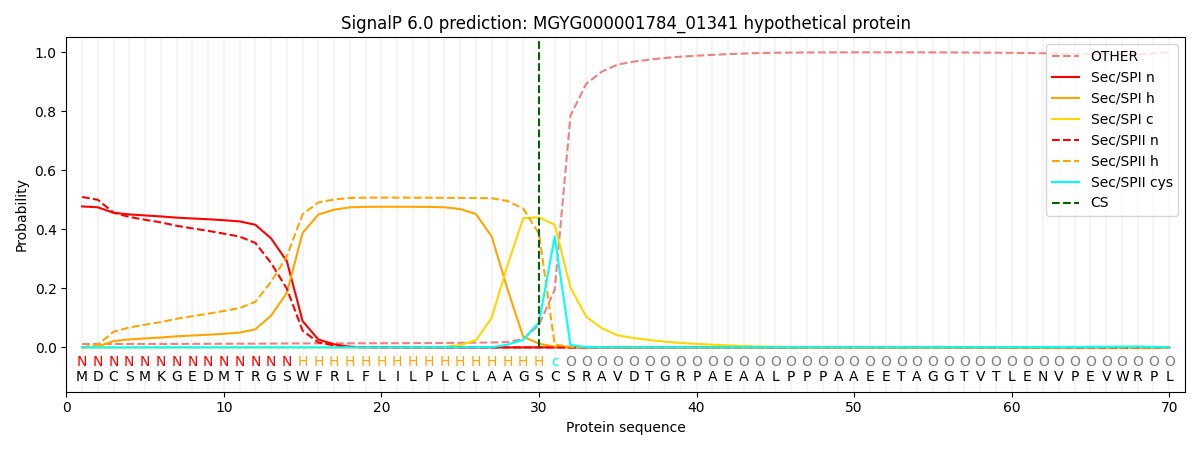
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014680 | 0.458243 | 0.522773 | 0.002210 | 0.001473 | 0.000586 |
