You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000004207_00026
You are here: Home > Sequence: MGYG000004207_00026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA6398 sp003150315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA6398; UBA6398 sp003150315 | |||||||||||
| CAZyme ID | MGYG000004207_00026 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 33664; End: 36822 Strand: - | |||||||||||
Full Sequence Download help
| MKLKNIAKTV ALSGISFLAA SNACAQFKQA SINTMESTFR CIPDSQKIAV YWYWMSDNIS | 60 |
| KEGVVKDLQA MKRVGINRVQ IGNIGSNDIA YGKVKFYSPE WWEVLHTALK TAGDLGIEVG | 120 |
| IFNSPGWSQS GGPWVKPNQA MRYLAESRTN VTGGKLLKIK LPEVGKEAED VKVLAFPDLE | 180 |
| TPTSFKAQQE IGSAKTIDFH CDKPATVRSI TFECKGNTFL NTAALYAKID NEYKFIRNIT | 240 |
| LDRRNAELNV GFVPFAPIAA SVPETTSSDF RLVFNGDQDK FKNITLSENP VVDSYAEKTL | 300 |
| EKMWQTPHPF WYDYMWKPSS ECSDAKLVMK TNQVIDLTEN FKDGVLTWNA PKGKWVILRT | 360 |
| AMRPTGTTNA PATPEATGYE TDKMSKKHIA SHFDAFLGEI MRRIPPQDRK TFKIVVEDSY | 420 |
| ETGGQNWTDE MIPEFKKRYG YDPVPFIPTF YGYVVNSEDQ SDRFLWDVRR LIADMVSYEY | 480 |
| VGGLKKISNE HGLTTWLECY GHWGFPGEFL QYGGQSDEIA GEFWSAGDLG DIENRIASSC | 540 |
| GHIYGKRRVW AESFTCGGPD FSRYPGEMKQ RGDKFFAEGI NNTLYHLYIQ QPDDNAPGIN | 600 |
| APFGNEFNRL NTWFSQLDVF NKYIKRCNYM LQQGYYVADV AYYIGEDAPK MTGIRNPEIP | 660 |
| QGYSYDFINA EVIEKSGRVE NGQLAIDGGM KYRVLVLPPQ NTMRPEFLAK LQELVKNGLT | 720 |
| IIGPKPEKSP SLQNYPEADK KVSEIATKMW GGEKYFNRVI QYGKGKVYPQ ASIEEVMADM | 780 |
| NVVPDFKSND PTLPITFLHR SLPDADVYFV ANQGDKAISF NGTFRAAGKT PELWNPQTAE | 840 |
| VRALPQFTRL SKTVEVPMKL EPLESAFIVF RNGSEQPSAN SVNYPEKKVL TTITTPWKVA | 900 |
| FEQGKGGPES TQTFETLTDW SKNADNKIKY FAGHAIYTNT FKISKLPEGE TYIDLGKVMV | 960 |
| MAKVKINGEE VGGVWTPPYR LNITKYLKKG VNTLEVDVVN NWRNRLIGEK NMPESERFTK | 1020 |
| QSFTYLNKDS ELQESGLIGP VEIQNYSYEL KK | 1052 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 330 | 1044 | 2.7e-236 | 0.8847087378640777 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 43 | 844 | 4 | 869 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO69761.1 | 0.0 | 1 | 1051 | 1 | 1049 |
| ANI89646.1 | 0.0 | 21 | 1052 | 14 | 1044 |
| QUT78107.1 | 0.0 | 3 | 1052 | 1 | 1049 |
| QDM11648.1 | 0.0 | 3 | 1052 | 1 | 1049 |
| QUT32845.1 | 0.0 | 3 | 1050 | 1 | 1047 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 2.00e-140 | 34 | 1043 | 41 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.28e-126 | 18 | 1041 | 14 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.44e-126 | 18 | 1041 | 14 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.53e-134 | 17 | 1043 | 12 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
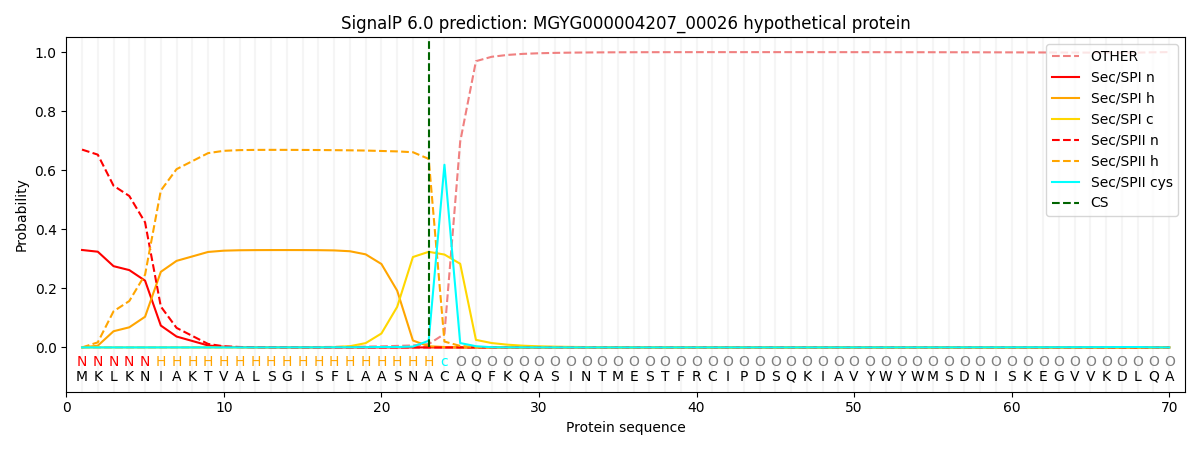
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000689 | 0.322741 | 0.676075 | 0.000201 | 0.000146 | 0.000136 |
